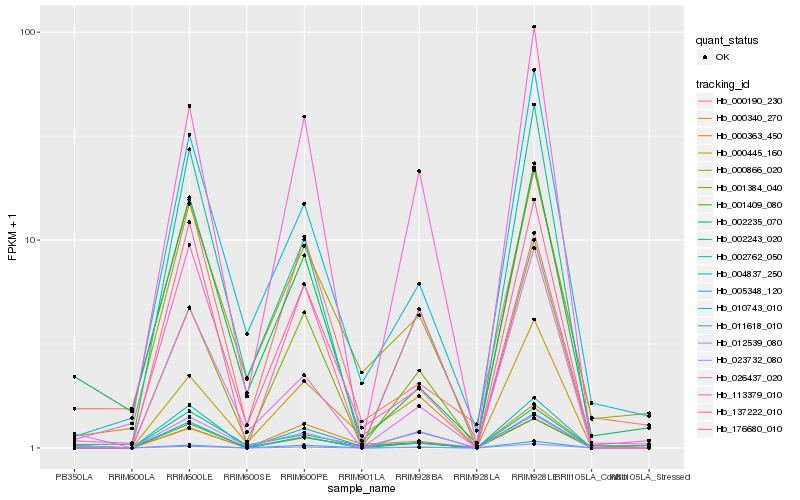
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000340_270 |
0.0 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 2 |
Hb_010743_010 |
0.1554394613 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 3 |
Hb_004837_250 |
0.1645523239 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_137222_010 |
0.1736997127 |
- |
- |
PREDICTED: hippocampus abundant transcript-like protein 1 [Jatropha curcas] |
| 5 |
Hb_001409_080 |
0.1760881453 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X1 [Populus euphratica] |
| 6 |
Hb_176680_010 |
0.1770770903 |
- |
- |
hypothetical protein JCGZ_22787 [Jatropha curcas] |
| 7 |
Hb_026437_020 |
0.1775355974 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 8 |
Hb_002235_070 |
0.1819661606 |
- |
- |
PREDICTED: uncharacterized protein LOC105644108 [Jatropha curcas] |
| 9 |
Hb_000363_450 |
0.1833077086 |
- |
- |
PREDICTED: phosphatidate cytidylyltransferase 1 [Jatropha curcas] |
| 10 |
Hb_000190_230 |
0.1897089667 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 11 |
Hb_002243_020 |
0.1898015529 |
- |
- |
PREDICTED: uncharacterized protein LOC105642097 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000866_020 |
0.1904366036 |
- |
- |
hypothetical protein B456_004G042000 [Gossypium raimondii] |
| 13 |
Hb_002762_050 |
0.1909930815 |
- |
- |
PREDICTED: uncharacterized protein LOC105640359 [Jatropha curcas] |
| 14 |
Hb_001384_040 |
0.1927320991 |
- |
- |
PREDICTED: beta-glucosidase 17-like [Jatropha curcas] |
| 15 |
Hb_011618_010 |
0.1928588725 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g59700 [Jatropha curcas] |
| 16 |
Hb_012539_080 |
0.1941071374 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 17 |
Hb_023732_080 |
0.1948830521 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 18 |
Hb_000445_160 |
0.1953815246 |
- |
- |
PREDICTED: probable calcium-binding protein CML44 [Jatropha curcas] |
| 19 |
Hb_113379_010 |
0.1955883314 |
- |
- |
PREDICTED: beta-xylosidase/alpha-L-arabinofuranosidase 1 [Jatropha curcas] |
| 20 |
Hb_005348_120 |
0.1959127004 |
- |
- |
PREDICTED: chlorophyll a-b binding protein, chloroplastic [Jatropha curcas] |