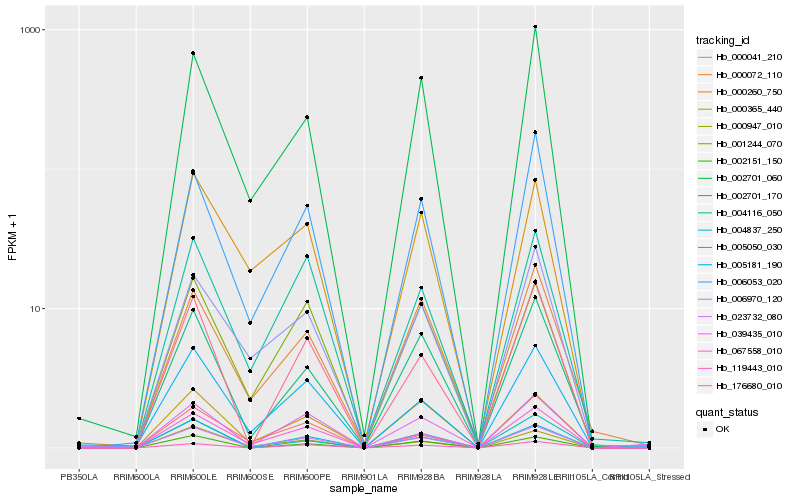
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_023732_080 |
0.0 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 2 |
Hb_005181_190 |
0.1068117654 |
- |
- |
PREDICTED: uncharacterized protein LOC105643550 [Jatropha curcas] |
| 3 |
Hb_067558_010 |
0.1213955422 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 4 |
Hb_005050_030 |
0.1256686055 |
- |
- |
PREDICTED: wall-associated receptor kinase 2-like [Jatropha curcas] |
| 5 |
Hb_000947_010 |
0.1317950271 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 6 |
Hb_004116_050 |
0.1319673581 |
- |
- |
PREDICTED: cytochrome P450 90A1 [Jatropha curcas] |
| 7 |
Hb_000365_440 |
0.134936543 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 8 |
Hb_001244_070 |
0.1352254052 |
- |
- |
hypothetical protein POPTR_0004s13470g [Populus trichocarpa] |
| 9 |
Hb_002701_060 |
0.1360295992 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002701_170 |
0.1366560465 |
- |
- |
PREDICTED: cytochrome P450 85A [Jatropha curcas] |
| 11 |
Hb_006053_020 |
0.140322058 |
- |
- |
CAP [Theobroma cacao] |
| 12 |
Hb_000260_750 |
0.1474552603 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 7-like [Jatropha curcas] |
| 13 |
Hb_004837_250 |
0.148567089 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002151_150 |
0.1488283058 |
- |
- |
PREDICTED: uncharacterized protein LOC102707106 [Oryza brachyantha] |
| 15 |
Hb_039435_010 |
0.1489759189 |
- |
- |
hypothetical protein SORBIDRAFT_10g025530 [Sorghum bicolor] |
| 16 |
Hb_006970_120 |
0.1500015119 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000041_210 |
0.150260278 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 42 [Jatropha curcas] |
| 18 |
Hb_176680_010 |
0.1518223121 |
- |
- |
hypothetical protein JCGZ_22787 [Jatropha curcas] |
| 19 |
Hb_119443_010 |
0.1526946776 |
- |
- |
hypothetical protein JCGZ_17348 [Jatropha curcas] |
| 20 |
Hb_000072_110 |
0.1552827645 |
- |
- |
PREDICTED: U-box domain-containing protein 35 [Jatropha curcas] |