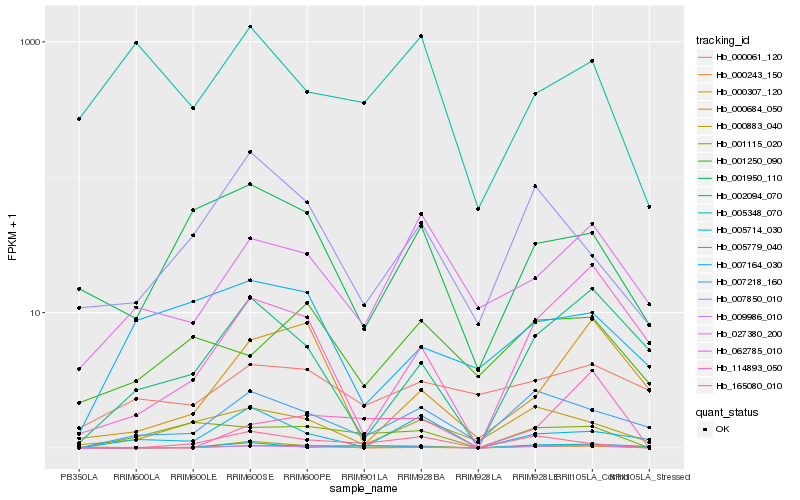
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009986_010 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 2 |
Hb_002094_070 |
0.2588993638 |
- |
- |
Organic cation transporter, putative [Ricinus communis] |
| 3 |
Hb_062785_010 |
0.2630330096 |
- |
- |
Organic cation transporter, putative [Ricinus communis] |
| 4 |
Hb_007218_160 |
0.2654941753 |
- |
- |
PREDICTED: uncharacterized protein LOC105108358 [Populus euphratica] |
| 5 |
Hb_001115_020 |
0.2697587805 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 2-like [Populus euphratica] |
| 6 |
Hb_000684_050 |
0.2982814934 |
- |
- |
PREDICTED: MLO-like protein 6 [Sesamum indicum] |
| 7 |
Hb_000243_150 |
0.3016258989 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_027380_200 |
0.3125502888 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 6 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000883_040 |
0.3138847121 |
- |
- |
- |
| 10 |
Hb_165080_010 |
0.3266417529 |
- |
- |
hypothetical protein JCGZ_01613 [Jatropha curcas] |
| 11 |
Hb_114893_050 |
0.3297621037 |
- |
- |
Uncharacterized protein isoform 3 [Theobroma cacao] |
| 12 |
Hb_000307_120 |
0.3301662628 |
- |
- |
hypothetical protein B456_002G120900 [Gossypium raimondii] |
| 13 |
Hb_005779_040 |
0.3355173176 |
- |
- |
- |
| 14 |
Hb_000061_120 |
0.3355268176 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 16 [Jatropha curcas] |
| 15 |
Hb_005348_070 |
0.3366128094 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_007850_010 |
0.3375678189 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.4 [Jatropha curcas] |
| 17 |
Hb_007164_030 |
0.3400148553 |
transcription factor |
TF Family: Orphans |
two-component system sensor histidine kinase/response regulator, putative [Ricinus communis] |
| 18 |
Hb_005714_030 |
0.340237732 |
- |
- |
PREDICTED: COBRA-like protein 6 isoform X1 [Gossypium raimondii] |
| 19 |
Hb_001950_110 |
0.3427284315 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB44 [Jatropha curcas] |
| 20 |
Hb_001250_090 |
0.3464478255 |
- |
- |
PREDICTED: protein NSP-INTERACTING KINASE 3 [Jatropha curcas] |