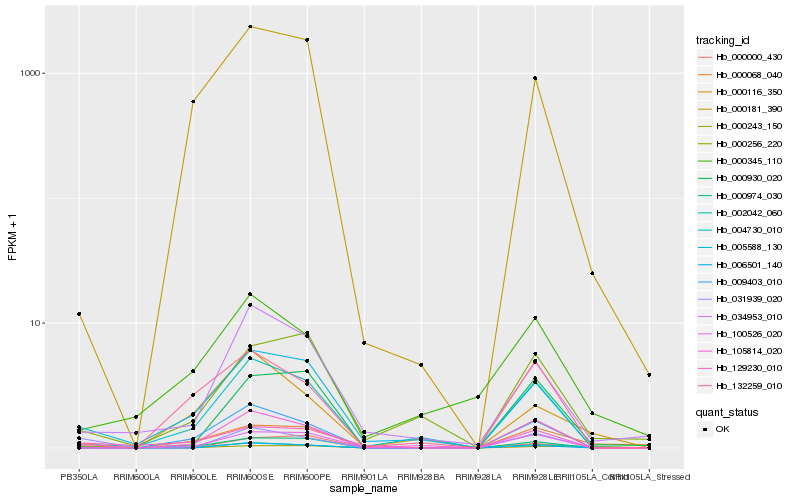
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002042_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_031939_020 |
0.2573359977 |
- |
- |
- |
| 3 |
Hb_005588_130 |
0.2650181503 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004730_010 |
0.2687312104 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT5 [Jatropha curcas] |
| 5 |
Hb_000068_040 |
0.2697377452 |
- |
- |
Inter-alpha-trypsin inhibitor heavy chain H3 precursor, putative [Ricinus communis] |
| 6 |
Hb_034953_010 |
0.2765086854 |
- |
- |
hypothetical protein JCGZ_00031 [Jatropha curcas] |
| 7 |
Hb_009403_010 |
0.2820109338 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 8 |
Hb_000116_350 |
0.2836462755 |
- |
- |
PREDICTED: uncharacterized protein LOC105628582 [Jatropha curcas] |
| 9 |
Hb_000930_020 |
0.2843876732 |
- |
- |
hypothetical protein PRUPE_ppa026493mg [Prunus persica] |
| 10 |
Hb_000256_220 |
0.2868941141 |
- |
- |
PREDICTED: flotillin-like protein 4 [Jatropha curcas] |
| 11 |
Hb_000345_110 |
0.2876649154 |
- |
- |
PREDICTED: uncharacterized protein LOC105647507 [Jatropha curcas] |
| 12 |
Hb_100526_020 |
0.2895036978 |
- |
- |
hypothetical protein POPTR_0003s09340g [Populus trichocarpa] |
| 13 |
Hb_000243_150 |
0.2925605209 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_000974_030 |
0.2930382035 |
- |
- |
PREDICTED: uncharacterized protein LOC105128463 isoform X1 [Populus euphratica] |
| 15 |
Hb_129230_010 |
0.2934342286 |
- |
- |
PREDICTED: inactive protein RESTRICTED TEV MOVEMENT 1-like [Nicotiana sylvestris] |
| 16 |
Hb_006501_140 |
0.2961976796 |
- |
- |
PREDICTED: MATE efflux family protein 9-like [Jatropha curcas] |
| 17 |
Hb_105814_020 |
0.2978524752 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |
| 18 |
Hb_000000_430 |
0.2989284793 |
- |
- |
hypothetical protein POPTR_0014s04130g [Populus trichocarpa] |
| 19 |
Hb_000181_390 |
0.2990786877 |
- |
- |
unknown [Lotus japonicus] |
| 20 |
Hb_132259_010 |
0.3038673737 |
- |
- |
PREDICTED: WAT1-related protein At5g64700 [Jatropha curcas] |