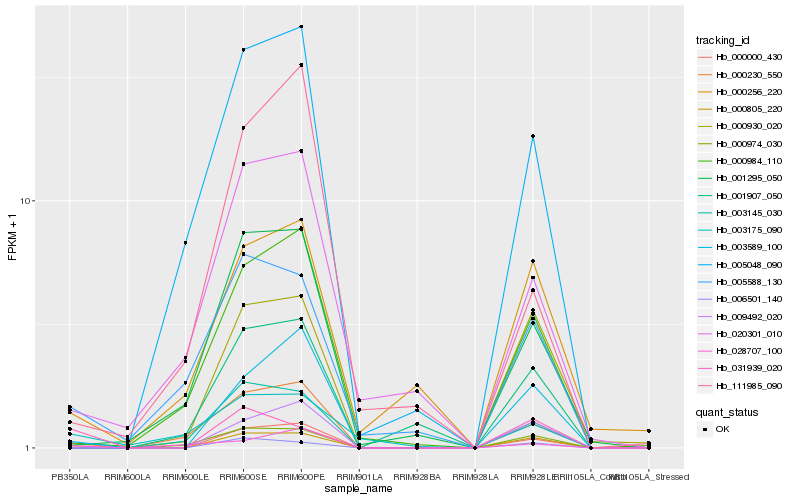
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000000_430 |
0.0 |
- |
- |
hypothetical protein POPTR_0014s04130g [Populus trichocarpa] |
| 2 |
Hb_000230_550 |
0.2171666893 |
- |
- |
PREDICTED: adenylate isopentenyltransferase 5, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000805_220 |
0.2238756638 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 4 |
Hb_009492_020 |
0.2259624798 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 5 |
Hb_003589_100 |
0.2266046273 |
- |
- |
hypothetical protein JCGZ_03338 [Jatropha curcas] |
| 6 |
Hb_000984_110 |
0.2310515996 |
- |
- |
hypothetical protein JCGZ_15953 [Jatropha curcas] |
| 7 |
Hb_001295_050 |
0.2427532559 |
- |
- |
PREDICTED: uncharacterized protein LOC101503108 isoform X1 [Cicer arietinum] |
| 8 |
Hb_020301_010 |
0.2475722513 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 9 |
Hb_005588_130 |
0.248593489 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003145_030 |
0.2505431468 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005048_090 |
0.2515171673 |
- |
- |
PREDICTED: glycine-rich cell wall structural protein 2 [Jatropha curcas] |
| 12 |
Hb_028707_100 |
0.2569839105 |
- |
- |
hypothetical protein RCOM_1687010 [Ricinus communis] |
| 13 |
Hb_031939_020 |
0.2605777215 |
- |
- |
- |
| 14 |
Hb_006501_140 |
0.2618421 |
- |
- |
PREDICTED: MATE efflux family protein 9-like [Jatropha curcas] |
| 15 |
Hb_111985_090 |
0.262304668 |
- |
- |
hypothetical protein RCOM_1595950 [Ricinus communis] |
| 16 |
Hb_000930_020 |
0.2642114078 |
- |
- |
hypothetical protein PRUPE_ppa026493mg [Prunus persica] |
| 17 |
Hb_000974_030 |
0.2669853516 |
- |
- |
PREDICTED: uncharacterized protein LOC105128463 isoform X1 [Populus euphratica] |
| 18 |
Hb_001907_050 |
0.2692212839 |
- |
- |
Receptor like protein 6, putative [Theobroma cacao] |
| 19 |
Hb_003175_090 |
0.2709787146 |
- |
- |
hypothetical protein POPTR_0011s01120g [Populus trichocarpa] |
| 20 |
Hb_000256_220 |
0.2720117777 |
- |
- |
PREDICTED: flotillin-like protein 4 [Jatropha curcas] |