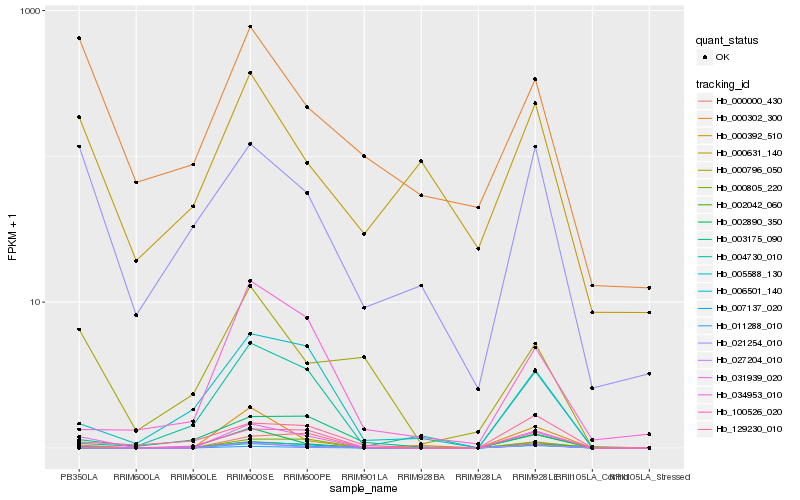
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031939_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_011288_010 |
0.1903884254 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 3 |
Hb_002042_060 |
0.2573359977 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000000_430 |
0.2605777215 |
- |
- |
hypothetical protein POPTR_0014s04130g [Populus trichocarpa] |
| 5 |
Hb_027204_010 |
0.2780646984 |
- |
- |
- |
| 6 |
Hb_021254_010 |
0.299062143 |
transcription factor |
TF Family: HSF |
hypothetical protein JCGZ_07655 [Jatropha curcas] |
| 7 |
Hb_000392_510 |
0.3031592613 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 36 kDa-like [Jatropha curcas] |
| 8 |
Hb_006501_140 |
0.3081889367 |
- |
- |
PREDICTED: MATE efflux family protein 9-like [Jatropha curcas] |
| 9 |
Hb_129230_010 |
0.3094327097 |
- |
- |
PREDICTED: inactive protein RESTRICTED TEV MOVEMENT 1-like [Nicotiana sylvestris] |
| 10 |
Hb_005588_130 |
0.3110144091 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004730_010 |
0.3235716008 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT5 [Jatropha curcas] |
| 12 |
Hb_000805_220 |
0.3309893959 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 13 |
Hb_000796_050 |
0.3319770856 |
- |
- |
galactinol synthase [Manihot esculenta] |
| 14 |
Hb_034953_010 |
0.3360394317 |
- |
- |
hypothetical protein JCGZ_00031 [Jatropha curcas] |
| 15 |
Hb_000302_300 |
0.3365611782 |
- |
- |
PREDICTED: heat shock protein 83 [Populus euphratica] |
| 16 |
Hb_007137_020 |
0.3390153765 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 17 |
Hb_000631_140 |
0.3415306771 |
- |
- |
PREDICTED: chaperone protein ClpB1 [Jatropha curcas] |
| 18 |
Hb_002890_350 |
0.3437575496 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 19 |
Hb_100526_020 |
0.3440491715 |
- |
- |
hypothetical protein POPTR_0003s09340g [Populus trichocarpa] |
| 20 |
Hb_003175_090 |
0.3447910206 |
- |
- |
hypothetical protein POPTR_0011s01120g [Populus trichocarpa] |