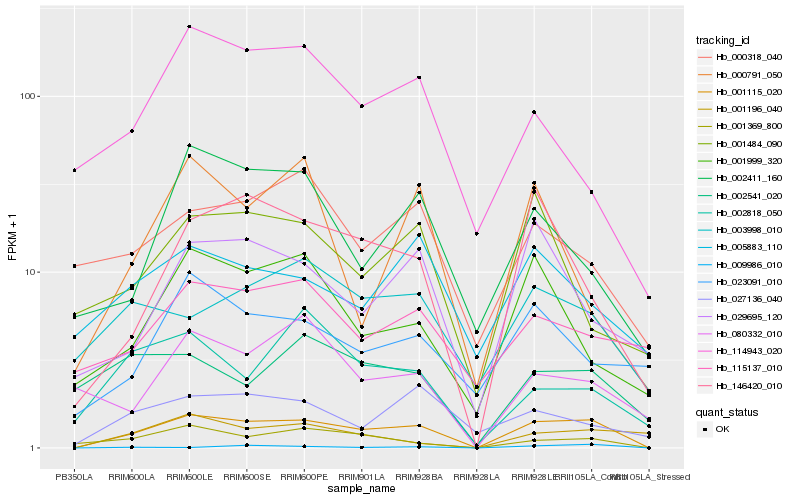
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001115_020 |
0.0 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 2-like [Populus euphratica] |
| 2 |
Hb_146420_010 |
0.2207884922 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_001369_800 |
0.234317333 |
- |
- |
hypothetical protein B456_011G005000 [Gossypium raimondii] |
| 4 |
Hb_029695_120 |
0.2513920898 |
- |
- |
hypothetical protein POPTR_0002s05320g [Populus trichocarpa] |
| 5 |
Hb_003998_010 |
0.2543460525 |
- |
- |
PREDICTED: protein BRASSINOSTEROID INSENSITIVE 1 [Jatropha curcas] |
| 6 |
Hb_001999_320 |
0.2558139077 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA [Jatropha curcas] |
| 7 |
Hb_002411_160 |
0.2564905542 |
- |
- |
PREDICTED: uncharacterized protein LOC105631262 isoform X1 [Jatropha curcas] |
| 8 |
Hb_027136_040 |
0.2587859633 |
- |
- |
hypothetical protein JCGZ_00277 [Jatropha curcas] |
| 9 |
Hb_000318_040 |
0.2649152251 |
- |
- |
hypothetical protein JCGZ_04877 [Jatropha curcas] |
| 10 |
Hb_023091_010 |
0.266434466 |
- |
- |
hypothetical protein POPTR_0016s06670g [Populus trichocarpa] |
| 11 |
Hb_005883_110 |
0.2679479033 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH79-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_002541_020 |
0.2686382501 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 13 |
Hb_001484_090 |
0.2688487166 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 14 |
Hb_114943_020 |
0.2689572112 |
- |
- |
PREDICTED: NADPH--cytochrome P450 reductase 2 [Jatropha curcas] |
| 15 |
Hb_002818_050 |
0.26914418 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 16 |
Hb_080332_010 |
0.2697269237 |
- |
- |
hypothetical protein POPTR_0018s10590g [Populus trichocarpa] |
| 17 |
Hb_009986_010 |
0.2697587805 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 18 |
Hb_001196_040 |
0.2717857319 |
- |
- |
PREDICTED: uncharacterized protein LOC104100193 [Nicotiana tomentosiformis] |
| 19 |
Hb_115137_010 |
0.2718003834 |
- |
- |
PREDICTED: probable protein S-acyltransferase 23 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000791_050 |
0.2734470244 |
- |
- |
PREDICTED: uncharacterized protein LOC105636760 [Jatropha curcas] |