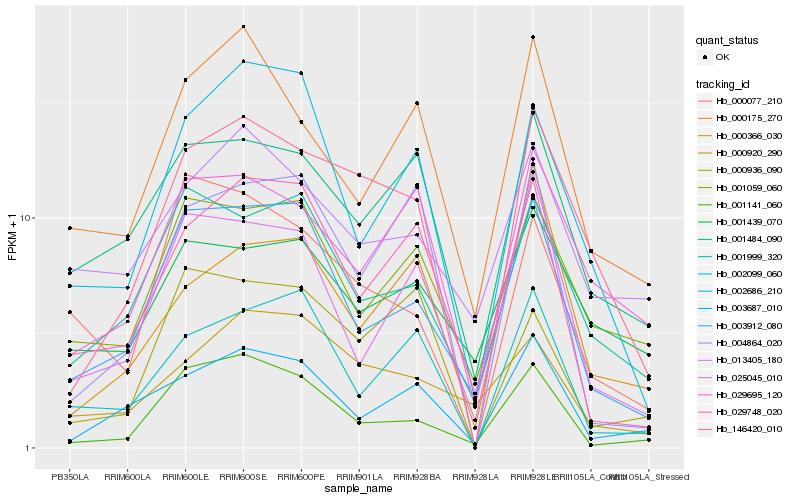
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_146420_010 |
0.0 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_029695_120 |
0.1588886171 |
- |
- |
hypothetical protein POPTR_0002s05320g [Populus trichocarpa] |
| 3 |
Hb_001999_320 |
0.1757398276 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA [Jatropha curcas] |
| 4 |
Hb_001484_090 |
0.1768363181 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 5 |
Hb_003687_010 |
0.1840882518 |
- |
- |
Cysteine-rich RLK 29, putative [Theobroma cacao] |
| 6 |
Hb_003912_080 |
0.1851932377 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 7 |
Hb_002686_210 |
0.1894435071 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_000175_270 |
0.1895462873 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001059_060 |
0.1915644588 |
- |
- |
PREDICTED: bifunctional riboflavin kinase/FMN phosphatase [Jatropha curcas] |
| 10 |
Hb_004864_020 |
0.1942271283 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 11 |
Hb_000366_030 |
0.1949130444 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Jatropha curcas] |
| 12 |
Hb_001141_060 |
0.1981828504 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_025045_010 |
0.1985518094 |
- |
- |
catalytic, putative [Ricinus communis] |
| 14 |
Hb_000936_090 |
0.2007714179 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RING1a isoform X2 [Jatropha curcas] |
| 15 |
Hb_029748_020 |
0.2009410177 |
- |
- |
PREDICTED: ent-kaur-16-ene synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_001439_070 |
0.2011987978 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 17 |
Hb_000077_210 |
0.2041301625 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1 [Jatropha curcas] |
| 18 |
Hb_002099_060 |
0.2044604592 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 19 |
Hb_013405_180 |
0.2051347903 |
- |
- |
PREDICTED: uncharacterized protein LOC105648441 [Jatropha curcas] |
| 20 |
Hb_000920_290 |
0.206955194 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |