| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
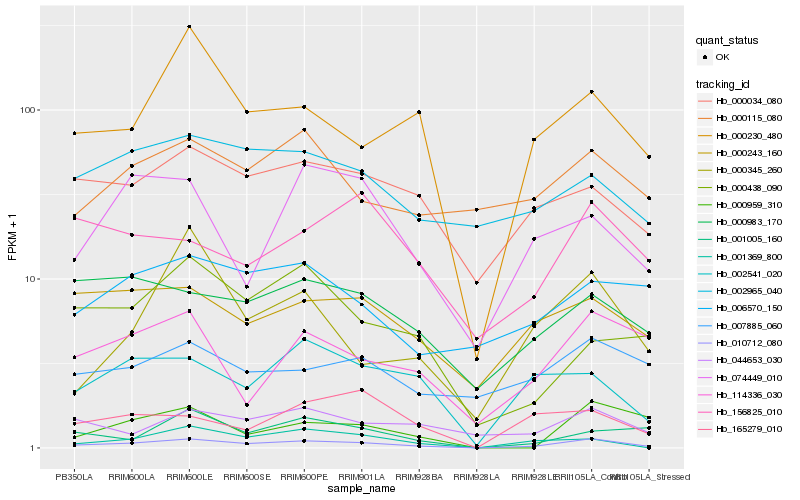
Hb_010712_080 |
0.0 |
- |
- |
hypothetical protein AALP_AA5G019400 [Arabis alpina] |
| 2 |
Hb_000230_480 |
0.2026393082 |
- |
- |
PREDICTED: endo-1,3;1,4-beta-D-glucanase-like [Jatropha curcas] |
| 3 |
Hb_002541_020 |
0.2041150904 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 4 |
Hb_001369_800 |
0.2077027486 |
- |
- |
hypothetical protein B456_011G005000 [Gossypium raimondii] |
| 5 |
Hb_000034_080 |
0.2079444188 |
- |
- |
PREDICTED: MADS-box protein AGL24-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_114336_030 |
0.2134871266 |
- |
- |
PREDICTED: uncharacterized protein LOC105642104 [Jatropha curcas] |
| 7 |
Hb_000243_160 |
0.2136911823 |
- |
- |
glutaredoxin-1, grx1, putative [Ricinus communis] |
| 8 |
Hb_000983_170 |
0.2142074554 |
- |
- |
PREDICTED: inositol-pentakisphosphate 2-kinase-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_044653_030 |
0.2156857781 |
- |
- |
- |
| 10 |
Hb_074449_010 |
0.2167716671 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 11 |
Hb_001005_160 |
0.2183810385 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002965_040 |
0.2188347026 |
- |
- |
drought-responsive family protein [Populus trichocarpa] |
| 13 |
Hb_006570_150 |
0.2231189028 |
- |
- |
PREDICTED: uncharacterized protein LOC105635342 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000115_080 |
0.2234109163 |
- |
- |
PREDICTED: uncharacterized protein LOC105642032 [Jatropha curcas] |
| 15 |
Hb_000345_260 |
0.2234767431 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Citrus sinensis] |
| 16 |
Hb_000959_310 |
0.2242313532 |
- |
- |
PREDICTED: WD repeat domain-containing protein 83 [Jatropha curcas] |
| 17 |
Hb_000438_090 |
0.2255360592 |
- |
- |
PREDICTED: mini-chromosome maintenance complex-binding protein [Jatropha curcas] |
| 18 |
Hb_007885_060 |
0.2277621925 |
- |
- |
replication factor C 40 kDa family protein [Populus trichocarpa] |
| 19 |
Hb_165279_010 |
0.2282928472 |
- |
- |
Kiwellin, putative [Ricinus communis] |
| 20 |
Hb_156825_010 |
0.2288903135 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein JCGZ_05054 [Jatropha curcas] |