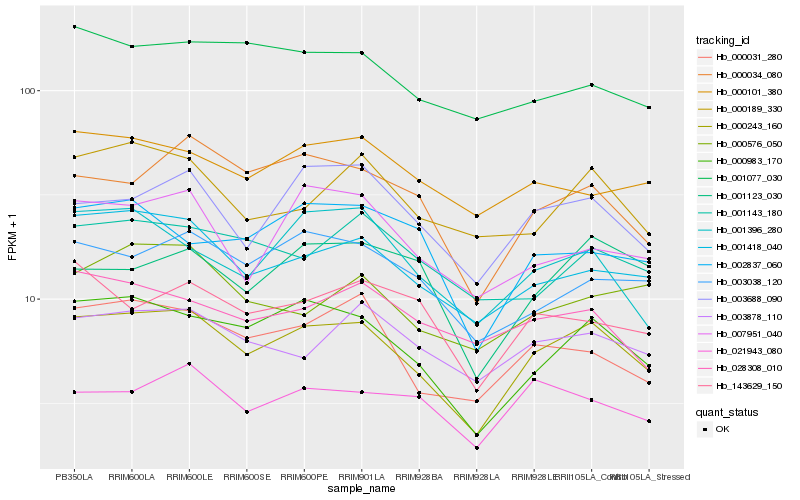
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000243_160 |
0.0 |
- |
- |
glutaredoxin-1, grx1, putative [Ricinus communis] |
| 2 |
Hb_000983_170 |
0.0710186409 |
- |
- |
PREDICTED: inositol-pentakisphosphate 2-kinase-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_001418_040 |
0.0879532053 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1-like [Jatropha curcas] |
| 4 |
Hb_000031_280 |
0.095088826 |
- |
- |
- |
| 5 |
Hb_000034_080 |
0.0961982578 |
- |
- |
PREDICTED: MADS-box protein AGL24-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_003688_090 |
0.0984309655 |
- |
- |
PREDICTED: ATP synthase subunit gamma, mitochondrial [Jatropha curcas] |
| 7 |
Hb_003038_120 |
0.1020459656 |
- |
- |
rer1 protein, putative [Ricinus communis] |
| 8 |
Hb_003878_110 |
0.103875683 |
- |
- |
PREDICTED: uncharacterized protein LOC105637024 [Jatropha curcas] |
| 9 |
Hb_000189_330 |
0.1053227682 |
- |
- |
PREDICTED: ER membrane protein complex subunit 2 [Jatropha curcas] |
| 10 |
Hb_001396_280 |
0.1082974831 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 2 [Jatropha curcas] |
| 11 |
Hb_007951_040 |
0.1087455019 |
- |
- |
signal peptidase family protein [Populus trichocarpa] |
| 12 |
Hb_021943_080 |
0.1092512417 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_143629_150 |
0.1120546366 |
- |
- |
PREDICTED: flap endonuclease 1 isoform X2 [Prunus mume] |
| 14 |
Hb_001143_180 |
0.1130305456 |
- |
- |
PREDICTED: replication factor C subunit 5 [Jatropha curcas] |
| 15 |
Hb_000101_380 |
0.1132475146 |
- |
- |
PREDICTED: clathrin light chain 2-like isoform X2 [Populus euphratica] |
| 16 |
Hb_001123_030 |
0.1132514617 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 17 |
Hb_028308_010 |
0.114018773 |
- |
- |
PREDICTED: probable protein S-acyltransferase 4 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001077_030 |
0.1140540113 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_002837_060 |
0.1142367371 |
- |
- |
PREDICTED: uncharacterized protein At1g10890-like [Nelumbo nucifera] |
| 20 |
Hb_000576_050 |
0.114269814 |
- |
- |
Kinase superfamily protein [Theobroma cacao] |