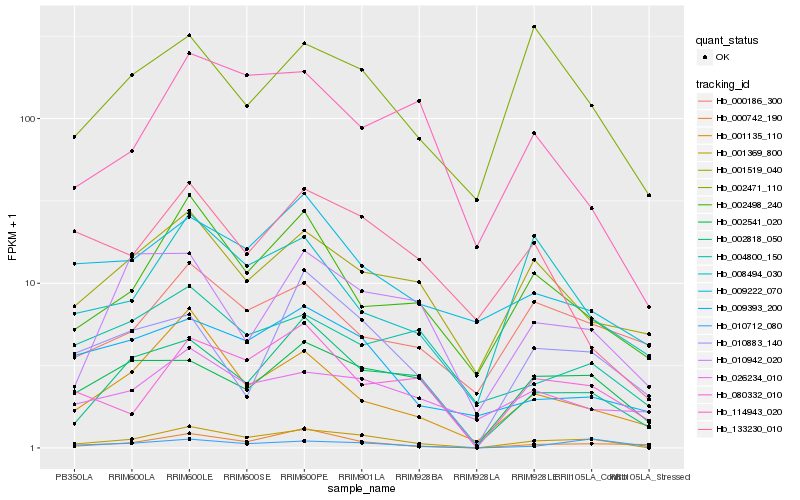
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001369_800 |
0.0 |
- |
- |
hypothetical protein B456_011G005000 [Gossypium raimondii] |
| 2 |
Hb_002818_050 |
0.1804745807 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 3 |
Hb_002541_020 |
0.2011619753 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 4 |
Hb_080332_010 |
0.2020973866 |
- |
- |
hypothetical protein POPTR_0018s10590g [Populus trichocarpa] |
| 5 |
Hb_002498_240 |
0.2042519813 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_010712_080 |
0.2077027486 |
- |
- |
hypothetical protein AALP_AA5G019400 [Arabis alpina] |
| 7 |
Hb_001135_110 |
0.2097074814 |
- |
- |
- |
| 8 |
Hb_000742_190 |
0.209783521 |
- |
- |
PREDICTED: histidinol dehydrogenase, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_001519_040 |
0.2121438026 |
- |
- |
PREDICTED: uncharacterized protein LOC105636007 [Jatropha curcas] |
| 10 |
Hb_009393_200 |
0.2214158702 |
- |
- |
PREDICTED: F-box only protein 13 [Jatropha curcas] |
| 11 |
Hb_114943_020 |
0.2222494482 |
- |
- |
PREDICTED: NADPH--cytochrome P450 reductase 2 [Jatropha curcas] |
| 12 |
Hb_010942_020 |
0.2263830411 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_009222_070 |
0.2269446037 |
- |
- |
PREDICTED: xylosyltransferase 2 [Jatropha curcas] |
| 14 |
Hb_010883_140 |
0.2278825744 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_002471_110 |
0.2287410327 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 28 [Jatropha curcas] |
| 16 |
Hb_008494_030 |
0.2291262873 |
- |
- |
catalytic, putative [Ricinus communis] |
| 17 |
Hb_133230_010 |
0.2306706936 |
- |
- |
hypothetical protein L484_001846 [Morus notabilis] |
| 18 |
Hb_026234_010 |
0.2329736561 |
- |
- |
choline monooxygenase, putative [Ricinus communis] |
| 19 |
Hb_000186_300 |
0.233275259 |
- |
- |
PREDICTED: malonyl-CoA decarboxylase, mitochondrial [Jatropha curcas] |
| 20 |
Hb_004800_150 |
0.2341466422 |
transcription factor |
TF Family: SNF2 |
PREDICTED: ATP-dependent DNA helicase DDM1 [Jatropha curcas] |