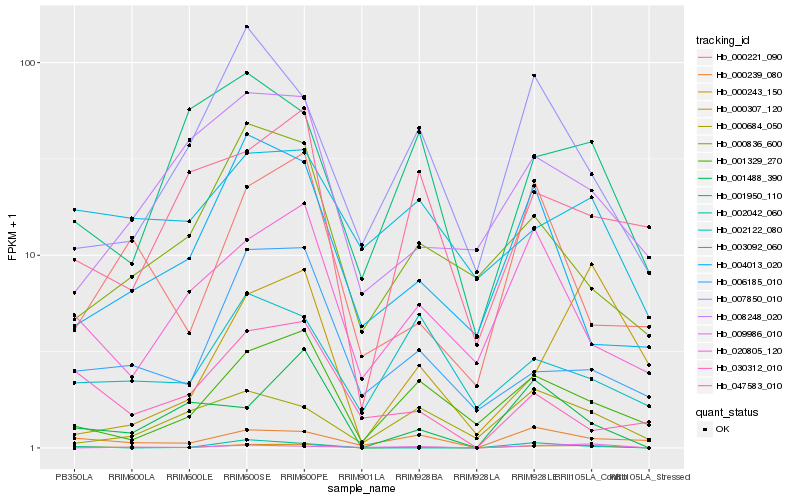
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000243_150 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_001329_270 |
0.278852094 |
- |
- |
PREDICTED: two-pore potassium channel 5 [Jatropha curcas] |
| 3 |
Hb_000239_080 |
0.2851392122 |
- |
- |
hypothetical protein POPTR_0014s01730g [Populus trichocarpa] |
| 4 |
Hb_001488_390 |
0.2869560584 |
- |
- |
SP1L, putative [Ricinus communis] |
| 5 |
Hb_002042_060 |
0.2925605209 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_020805_120 |
0.3004366005 |
- |
- |
PREDICTED: glutamate receptor 3.3 [Jatropha curcas] |
| 7 |
Hb_009986_010 |
0.3016258989 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 8 |
Hb_006185_010 |
0.301917186 |
- |
- |
hypothetical protein L484_013453 [Morus notabilis] |
| 9 |
Hb_001950_110 |
0.3077740834 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB44 [Jatropha curcas] |
| 10 |
Hb_007850_010 |
0.3094428593 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.4 [Jatropha curcas] |
| 11 |
Hb_003092_060 |
0.3104374877 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 12 |
Hb_000307_120 |
0.3106012072 |
- |
- |
hypothetical protein B456_002G120900 [Gossypium raimondii] |
| 13 |
Hb_000221_090 |
0.3133361805 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-6-like [Jatropha curcas] |
| 14 |
Hb_000836_600 |
0.3183290372 |
- |
- |
PREDICTED: delta(8)-fatty-acid desaturase 2 [Jatropha curcas] |
| 15 |
Hb_047583_010 |
0.3189137284 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 16 |
Hb_008248_020 |
0.3197698864 |
transcription factor |
TF Family: DBB |
zinc finger protein, putative [Ricinus communis] |
| 17 |
Hb_002122_080 |
0.322457976 |
- |
- |
- |
| 18 |
Hb_000684_050 |
0.322563565 |
- |
- |
PREDICTED: MLO-like protein 6 [Sesamum indicum] |
| 19 |
Hb_030312_010 |
0.3242167168 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004013_020 |
0.3244396775 |
- |
- |
PREDICTED: F-box protein SKIP2 [Jatropha curcas] |