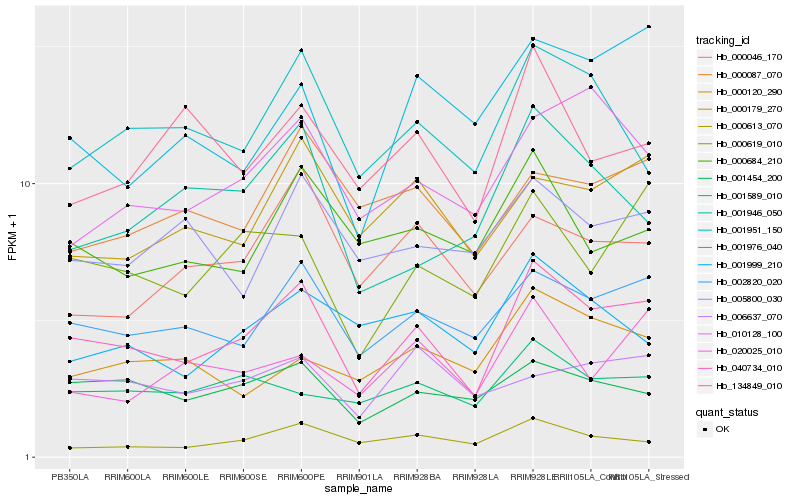
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_040734_010 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g37230 [Jatropha curcas] |
| 2 |
Hb_002820_020 |
0.1044659999 |
- |
- |
hypothetical protein VITISV_005587 [Vitis vinifera] |
| 3 |
Hb_000619_010 |
0.1257832114 |
- |
- |
TPA: hypothetical protein ZEAMMB73_187549 [Zea mays] |
| 4 |
Hb_001454_200 |
0.1312731227 |
transcription factor |
TF Family: TRAF |
PREDICTED: uncharacterized protein LOC105643434 [Jatropha curcas] |
| 5 |
Hb_001951_150 |
0.1384353228 |
- |
- |
PREDICTED: uncharacterized protein LOC105649758 [Jatropha curcas] |
| 6 |
Hb_000179_270 |
0.1386721314 |
- |
- |
hypothetical protein JCGZ_07444 [Jatropha curcas] |
| 7 |
Hb_010128_100 |
0.1410474976 |
- |
- |
PREDICTED: probable L-cysteine desulfhydrase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000684_210 |
0.1414240584 |
- |
- |
PREDICTED: uncharacterized protein LOC105642177 [Jatropha curcas] |
| 9 |
Hb_001589_010 |
0.1422311809 |
- |
- |
PREDICTED: uncharacterized protein LOC105638192 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000613_070 |
0.1423192595 |
- |
- |
- |
| 11 |
Hb_134849_010 |
0.1479413154 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 12 |
Hb_001976_040 |
0.1479680533 |
- |
- |
PREDICTED: nicotinamidase 1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000087_070 |
0.1480598315 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 14 |
Hb_001999_210 |
0.1486101339 |
- |
- |
PREDICTED: sodium-dependent phosphate transport protein 1, chloroplastic [Eucalyptus grandis] |
| 15 |
Hb_001946_050 |
0.1496091969 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005800_030 |
0.1504349408 |
- |
- |
PREDICTED: lysM and putative peptidoglycan-binding domain-containing protein 4-like [Jatropha curcas] |
| 17 |
Hb_000046_170 |
0.1506115444 |
- |
- |
PREDICTED: uncharacterized protein LOC105631841 isoform X1 [Jatropha curcas] |
| 18 |
Hb_020025_010 |
0.1531862787 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Jatropha curcas] |
| 19 |
Hb_000120_290 |
0.1533311945 |
- |
- |
PREDICTED: N-acylphosphatidylethanolamine synthase isoform X1 [Jatropha curcas] |
| 20 |
Hb_006637_070 |
0.1536467872 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g37230 [Jatropha curcas] |