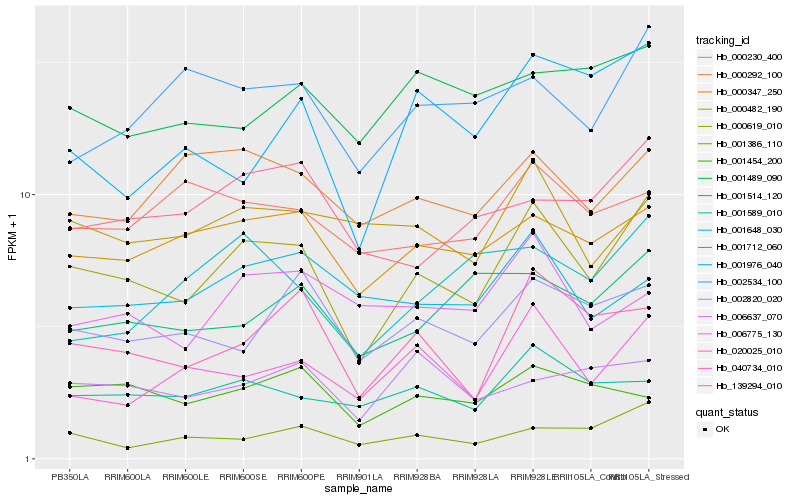
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000619_010 |
0.0 |
- |
- |
TPA: hypothetical protein ZEAMMB73_187549 [Zea mays] |
| 2 |
Hb_000347_250 |
0.1220016548 |
- |
- |
PREDICTED: uncharacterized protein LOC105133971 [Populus euphratica] |
| 3 |
Hb_040734_010 |
0.1257832114 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g37230 [Jatropha curcas] |
| 4 |
Hb_001589_010 |
0.1341123492 |
- |
- |
PREDICTED: uncharacterized protein LOC105638192 isoform X1 [Jatropha curcas] |
| 5 |
Hb_020025_010 |
0.1359900417 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Jatropha curcas] |
| 6 |
Hb_002820_020 |
0.1372057233 |
- |
- |
hypothetical protein VITISV_005587 [Vitis vinifera] |
| 7 |
Hb_139294_010 |
0.1403294169 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_002534_100 |
0.1420897585 |
- |
- |
PREDICTED: malate dehydrogenase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001514_120 |
0.1427310278 |
- |
- |
hypothetical protein VITISV_015004 [Vitis vinifera] |
| 10 |
Hb_000482_190 |
0.1429870007 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 11 |
Hb_000292_100 |
0.143225679 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1a [Jatropha curcas] |
| 12 |
Hb_001386_110 |
0.1454180725 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: putative fasciclin-like arabinogalactan protein 20 [Populus euphratica] |
| 13 |
Hb_001454_200 |
0.1483883138 |
transcription factor |
TF Family: TRAF |
PREDICTED: uncharacterized protein LOC105643434 [Jatropha curcas] |
| 14 |
Hb_000230_400 |
0.148708795 |
- |
- |
PREDICTED: protein ABCI12, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_001712_060 |
0.1503416579 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK9 [Jatropha curcas] |
| 16 |
Hb_001976_040 |
0.1509815645 |
- |
- |
PREDICTED: nicotinamidase 1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_001489_090 |
0.1530991177 |
- |
- |
PREDICTED: uncharacterized protein LOC105649775 [Jatropha curcas] |
| 18 |
Hb_006637_070 |
0.154031764 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g37230 [Jatropha curcas] |
| 19 |
Hb_001648_030 |
0.1542417439 |
- |
- |
PREDICTED: uncharacterized protein LOC105628931 isoform X2 [Jatropha curcas] |
| 20 |
Hb_006775_130 |
0.1547377578 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET2a-like [Jatropha curcas] |