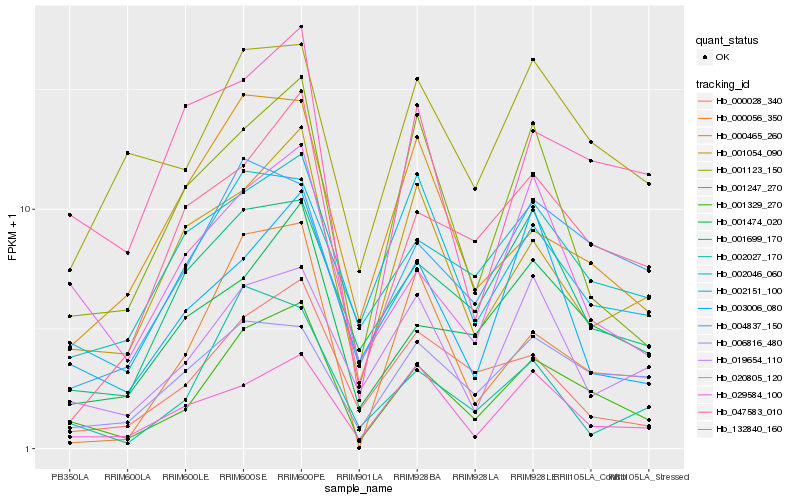
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001329_270 |
0.0 |
- |
- |
PREDICTED: two-pore potassium channel 5 [Jatropha curcas] |
| 2 |
Hb_001474_020 |
0.1578701297 |
- |
- |
PREDICTED: uncharacterized protein LOC105650917 [Jatropha curcas] |
| 3 |
Hb_001054_090 |
0.1662131993 |
- |
- |
Potassium channel SKOR, putative [Ricinus communis] |
| 4 |
Hb_132840_160 |
0.1686529193 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_020805_120 |
0.1688485787 |
- |
- |
PREDICTED: glutamate receptor 3.3 [Jatropha curcas] |
| 6 |
Hb_001247_270 |
0.1696659107 |
- |
- |
PREDICTED: probable L-cysteine desulfhydrase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_004837_150 |
0.1702324962 |
- |
- |
hypothetical protein POPTR_0001s26540g [Populus trichocarpa] |
| 8 |
Hb_000056_350 |
0.1733867924 |
- |
- |
PREDICTED: uncharacterized protein LOC105630437 [Jatropha curcas] |
| 9 |
Hb_019654_110 |
0.1739259192 |
- |
- |
PREDICTED: F-box protein SKIP23-like [Jatropha curcas] |
| 10 |
Hb_001699_170 |
0.1740479024 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000028_340 |
0.1746066637 |
- |
- |
PREDICTED: potassium channel SKOR-like [Prunus mume] |
| 12 |
Hb_002151_100 |
0.1746503592 |
- |
- |
PREDICTED: uncharacterized protein LOC105637951 [Jatropha curcas] |
| 13 |
Hb_029584_100 |
0.1755904788 |
- |
- |
Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase [Gossypium arboreum] |
| 14 |
Hb_002046_060 |
0.176082921 |
- |
- |
PREDICTED: beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 15 |
Hb_047583_010 |
0.1767116651 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 16 |
Hb_006816_480 |
0.1807411661 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001123_150 |
0.181358072 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 18 |
Hb_002027_170 |
0.1831516646 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RKF3 [Jatropha curcas] |
| 19 |
Hb_003006_080 |
0.1877268531 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000465_260 |
0.1889051064 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT4-like [Jatropha curcas] |