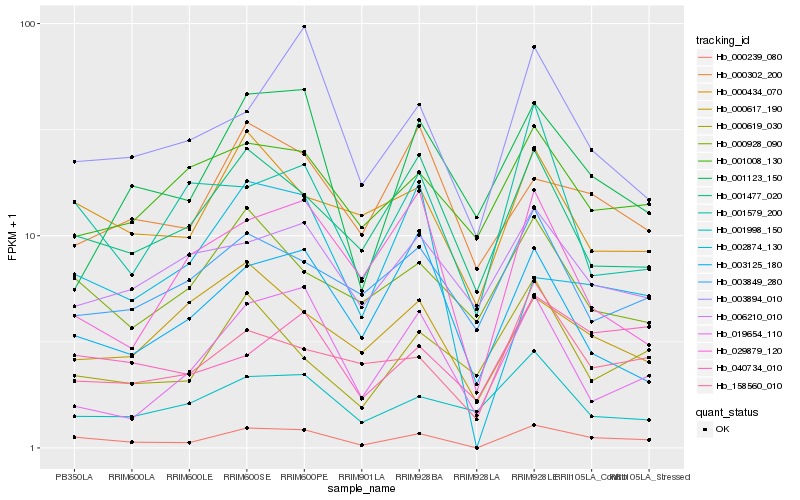
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000239_080 |
0.0 |
- |
- |
hypothetical protein POPTR_0014s01730g [Populus trichocarpa] |
| 2 |
Hb_000434_070 |
0.1947328472 |
- |
- |
PREDICTED: uncharacterized transmembrane protein DDB_G0289901 [Jatropha curcas] |
| 3 |
Hb_158560_010 |
0.1964624825 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable acyl-activating enzyme 1, peroxisomal [Jatropha curcas] |
| 4 |
Hb_001477_020 |
0.1969834052 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105646886 isoform X3 [Jatropha curcas] |
| 5 |
Hb_003894_010 |
0.2007848613 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_040734_010 |
0.2010152162 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g37230 [Jatropha curcas] |
| 7 |
Hb_001579_200 |
0.2077831818 |
- |
- |
nucleoredoxin, putative [Ricinus communis] |
| 8 |
Hb_002874_130 |
0.2126946028 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 9 [Jatropha curcas] |
| 9 |
Hb_019654_110 |
0.2127053147 |
- |
- |
PREDICTED: F-box protein SKIP23-like [Jatropha curcas] |
| 10 |
Hb_000928_090 |
0.2130075453 |
- |
- |
PREDICTED: septin and tuftelin-interacting protein 1 homolog 1 [Jatropha curcas] |
| 11 |
Hb_000617_190 |
0.2148389571 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase [Jatropha curcas] |
| 12 |
Hb_003125_180 |
0.2166464738 |
- |
- |
hypothetical protein JCGZ_26440 [Jatropha curcas] |
| 13 |
Hb_001123_150 |
0.2170054865 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 14 |
Hb_001998_150 |
0.2172091699 |
- |
- |
hypothetical protein JCGZ_09140 [Jatropha curcas] |
| 15 |
Hb_000302_200 |
0.2174316979 |
- |
- |
ornithine cyclodeaminase, putative [Ricinus communis] |
| 16 |
Hb_003849_280 |
0.217527894 |
- |
- |
PREDICTED: uncharacterized protein LOC105644977 [Jatropha curcas] |
| 17 |
Hb_000619_030 |
0.2179967262 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_006210_010 |
0.2191918508 |
- |
- |
PREDICTED: probable acyl-activating enzyme 17, peroxisomal isoform X2 [Jatropha curcas] |
| 19 |
Hb_029879_120 |
0.2199829242 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001008_130 |
0.2204332558 |
transcription factor |
TF Family: mTERF |
PREDICTED: cytochrome c1-2, heme protein, mitochondrial-like [Elaeis guineensis] |