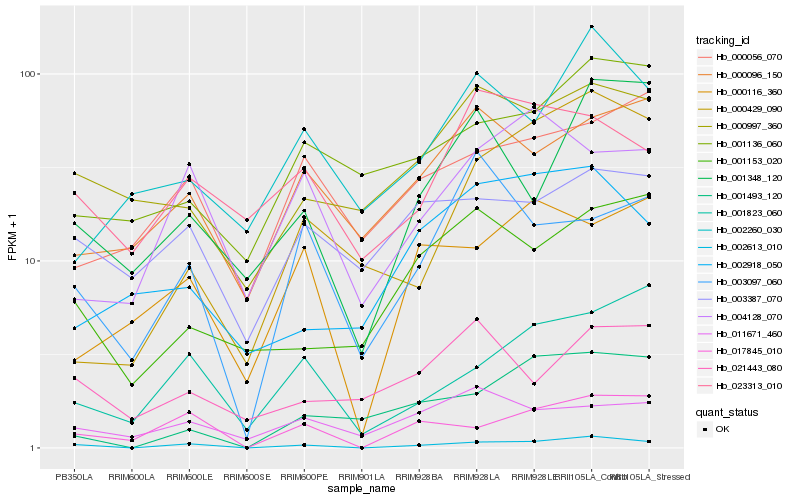
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002613_010 |
0.0 |
- |
- |
PREDICTED: copper chaperone for superoxide dismutase, chloroplastic/cytosolic [Jatropha curcas] |
| 2 |
Hb_017845_010 |
0.1887280176 |
- |
- |
- |
| 3 |
Hb_001823_060 |
0.2381020625 |
- |
- |
fructose-bisphosphate aldolase, putative [Ricinus communis] |
| 4 |
Hb_001348_120 |
0.2504874712 |
- |
- |
Phloem protein 2-B10, putative [Theobroma cacao] |
| 5 |
Hb_001493_120 |
0.256835971 |
- |
- |
- |
| 6 |
Hb_000997_360 |
0.2599837272 |
- |
- |
hypothetical protein AALP_AAs70636U000100 [Arabis alpina] |
| 7 |
Hb_003387_070 |
0.2632268317 |
- |
- |
Acyl-protein thioesterase, putative [Ricinus communis] |
| 8 |
Hb_004128_070 |
0.2656050146 |
- |
- |
PREDICTED: thioredoxin-like protein AAED1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_003097_060 |
0.266834243 |
- |
- |
PREDICTED: GEM-like protein 4 [Jatropha curcas] |
| 10 |
Hb_001153_020 |
0.2673679584 |
- |
- |
PREDICTED: histone deacetylase complex subunit SAP18-like [Cucumis melo] |
| 11 |
Hb_000116_360 |
0.2681040531 |
- |
- |
PREDICTED: uncharacterized protein LOC105116418 [Populus euphratica] |
| 12 |
Hb_000429_090 |
0.2681788023 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 3, chloroplastic [Jatropha curcas] |
| 13 |
Hb_023313_010 |
0.2701627113 |
- |
- |
PREDICTED: linolenate hydroperoxide lyase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000096_150 |
0.2713197894 |
- |
- |
NADH-ubiquinone oxidoreductase 18 kDa subunit, mitochondrial precursor, putative [Ricinus communis] |
| 15 |
Hb_001136_060 |
0.2739280262 |
- |
- |
PREDICTED: uncharacterized protein LOC105641994 [Jatropha curcas] |
| 16 |
Hb_002260_030 |
0.2743413825 |
- |
- |
PREDICTED: putative glutamine amidotransferase YLR126C [Jatropha curcas] |
| 17 |
Hb_011671_460 |
0.2753588965 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360 [Jatropha curcas] |
| 18 |
Hb_000056_070 |
0.2765257041 |
- |
- |
- |
| 19 |
Hb_002918_050 |
0.2779977903 |
transcription factor |
TF Family: SBP |
PREDICTED: mitochondrial arginine transporter BAC1-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_021443_080 |
0.2805706062 |
- |
- |
PREDICTED: CDGSH iron-sulfur domain-containing protein NEET [Jatropha curcas] |