| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
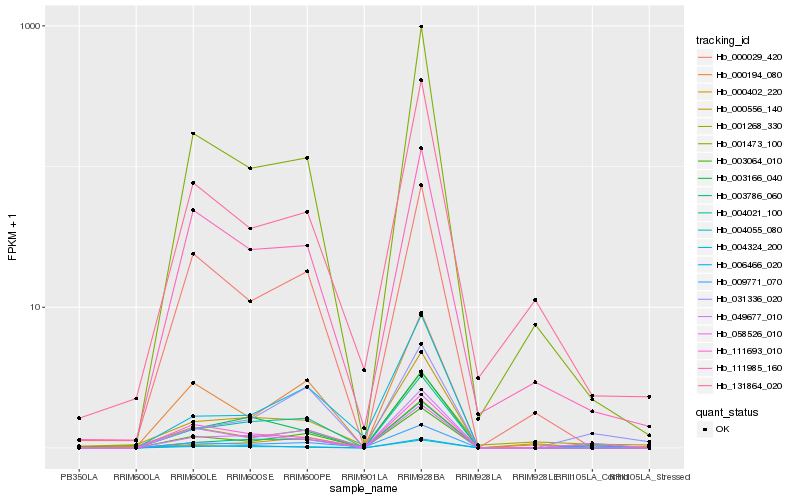
| 1 |
Hb_009771_070 |
0.0 |
- |
- |
hypothetical protein POPTR_0016s12140g [Populus trichocarpa] |
| 2 |
Hb_049677_010 |
0.1150445818 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 3 |
Hb_000402_220 |
0.1823488787 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 4 |
Hb_004324_200 |
0.1894311822 |
- |
- |
PREDICTED: transcription factor TRY-like [Populus euphratica] |
| 5 |
Hb_003064_010 |
0.1974189116 |
- |
- |
Senescence-related gene 1 [Theobroma cacao] |
| 6 |
Hb_031336_020 |
0.1984861074 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001473_100 |
0.2010377006 |
- |
- |
chalcone synthase 1 [Jatropha curcas] |
| 8 |
Hb_111985_160 |
0.2018663319 |
- |
- |
hypothetical protein B456_010G008900 [Gossypium raimondii] |
| 9 |
Hb_004055_080 |
0.2035636913 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 10 |
Hb_058526_010 |
0.2053355848 |
- |
- |
- |
| 11 |
Hb_131864_020 |
0.2074309982 |
- |
- |
PREDICTED: anthocyanidin reductase [Jatropha curcas] |
| 12 |
Hb_004021_100 |
0.2082349565 |
- |
- |
- |
| 13 |
Hb_000556_140 |
0.2103629414 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 9 [Jatropha curcas] |
| 14 |
Hb_000194_080 |
0.2110505228 |
- |
- |
hypothetical protein JCGZ_18743 [Jatropha curcas] |
| 15 |
Hb_000029_420 |
0.2118729662 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |
| 16 |
Hb_006466_020 |
0.2124489194 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Jatropha curcas] |
| 17 |
Hb_003786_060 |
0.2169201348 |
- |
- |
PREDICTED: transmembrane protein 220 [Jatropha curcas] |
| 18 |
Hb_111693_010 |
0.2179876105 |
- |
- |
hypothetical protein JCGZ_21650 [Jatropha curcas] |
| 19 |
Hb_001268_330 |
0.2224225759 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 20 |
Hb_003166_040 |
0.2236913421 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.1 [Jatropha curcas] |