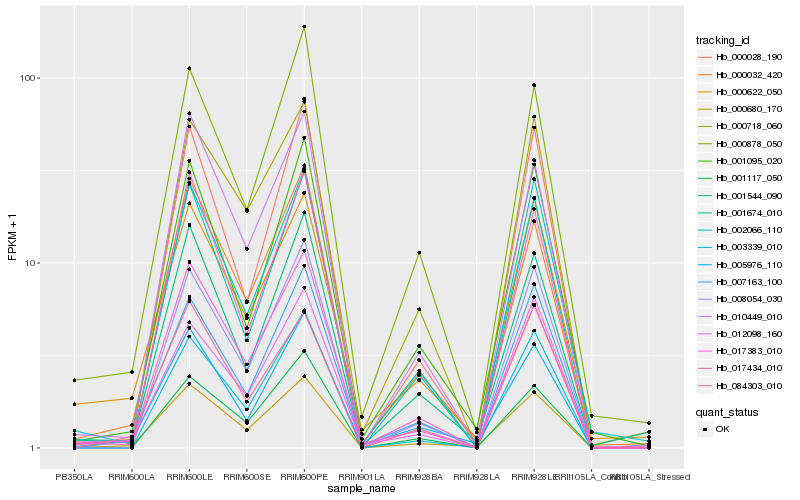
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000680_170 |
0.0 |
- |
- |
hypothetical protein JCGZ_24789 [Jatropha curcas] |
| 2 |
Hb_000032_420 |
0.0806785989 |
- |
- |
PREDICTED: GDSL esterase/lipase At4g10955 [Jatropha curcas] |
| 3 |
Hb_005976_110 |
0.086636687 |
- |
- |
PREDICTED: uncharacterized protein LOC105639708 [Jatropha curcas] |
| 4 |
Hb_017383_010 |
0.0879244414 |
- |
- |
PREDICTED: uncharacterized protein LOC105628176 [Jatropha curcas] |
| 5 |
Hb_010449_010 |
0.0915962184 |
- |
- |
l-ascorbate oxidase, putative [Ricinus communis] |
| 6 |
Hb_001674_010 |
0.0917515906 |
- |
- |
PREDICTED: reticulon-like protein B21 isoform X1 [Populus euphratica] |
| 7 |
Hb_001095_020 |
0.0969188977 |
- |
- |
PREDICTED: F-box protein At2g26850-like [Jatropha curcas] |
| 8 |
Hb_008054_030 |
0.0981540044 |
- |
- |
PREDICTED: VQ motif-containing protein 1-like [Jatropha curcas] |
| 9 |
Hb_001544_090 |
0.0982373663 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 10 |
Hb_002066_110 |
0.0993103413 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 11 |
Hb_000718_060 |
0.0998655577 |
transcription factor |
TF Family: MYB |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_007163_100 |
0.1005411026 |
- |
- |
BnaC03g52280D [Brassica napus] |
| 13 |
Hb_084303_010 |
0.1013375404 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000622_050 |
0.1020207091 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003339_010 |
0.1028945071 |
- |
- |
hypothetical protein JCGZ_04276 [Jatropha curcas] |
| 16 |
Hb_017434_010 |
0.1071017869 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 17 |
Hb_012098_160 |
0.1073095993 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 18 |
Hb_000878_050 |
0.1082603526 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 19 |
Hb_000028_190 |
0.1103972737 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g48480 [Jatropha curcas] |
| 20 |
Hb_001117_050 |
0.1113716803 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |