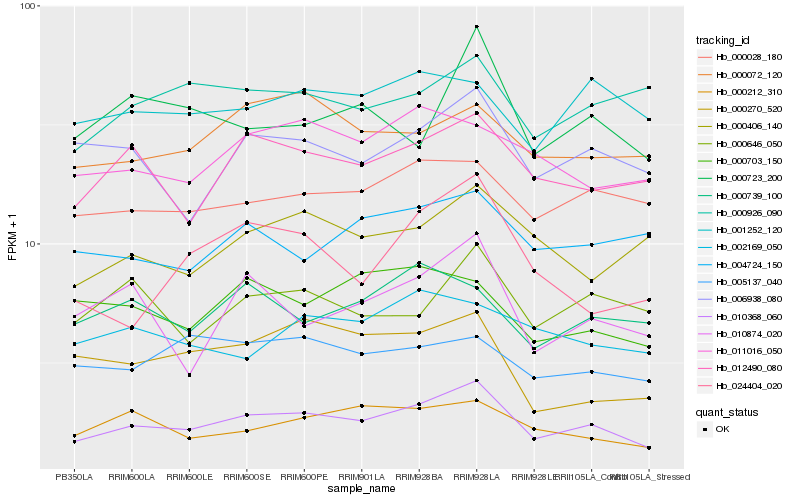
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010368_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_012490_080 |
0.1024413095 |
- |
- |
threonine synthase, putative [Ricinus communis] |
| 3 |
Hb_006938_080 |
0.1067675423 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 4 |
Hb_000028_180 |
0.107069083 |
- |
- |
PREDICTED: probable serine/threonine protein kinase IREH1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000212_310 |
0.1086590197 |
- |
- |
hypothetical protein JCGZ_24350 [Jatropha curcas] |
| 6 |
Hb_011016_050 |
0.1144473541 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI7 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000072_120 |
0.1155920262 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 8 |
Hb_010874_020 |
0.1174984294 |
- |
- |
PREDICTED: uncharacterized protein LOC105636745 [Jatropha curcas] |
| 9 |
Hb_000406_140 |
0.1176051159 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000646_050 |
0.1182528128 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 22-like [Jatropha curcas] |
| 11 |
Hb_001252_120 |
0.1191649809 |
- |
- |
PREDICTED: delta-1-pyrroline-5-carboxylate synthase-like [Gossypium raimondii] |
| 12 |
Hb_005137_040 |
0.1195062884 |
- |
- |
hypothetical protein JCGZ_12324 [Jatropha curcas] |
| 13 |
Hb_000270_520 |
0.119801854 |
- |
- |
PREDICTED: alpha-N-acetylglucosaminidase-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000723_200 |
0.1198589421 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105642236 isoform X1 [Jatropha curcas] |
| 15 |
Hb_024404_020 |
0.121237618 |
- |
- |
PREDICTED: putative F-box protein At1g65770 [Jatropha curcas] |
| 16 |
Hb_000703_150 |
0.1217226092 |
- |
- |
PREDICTED: F-box/LRR-repeat MAX2 homolog A [Jatropha curcas] |
| 17 |
Hb_000739_100 |
0.1219221137 |
- |
- |
ku P80 DNA helicase, putative [Ricinus communis] |
| 18 |
Hb_004724_150 |
0.1225975413 |
- |
- |
expressed protein, putative [Ricinus communis] |
| 19 |
Hb_000926_090 |
0.1226523956 |
- |
- |
PREDICTED: dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex 2, mitochondrial-like [Jatropha curcas] |
| 20 |
Hb_002169_050 |
0.1236495561 |
- |
- |
PREDICTED: inositol 1,3,4-trisphosphate 5/6-kinase 4 [Jatropha curcas] |