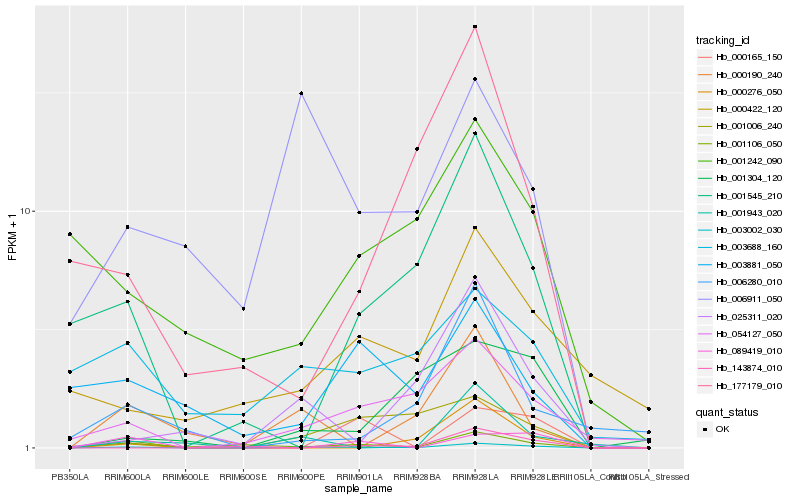
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003002_030 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 2 |
Hb_054127_050 |
0.2755343593 |
- |
- |
PREDICTED: uncharacterized protein LOC105638784 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001106_050 |
0.2954050442 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 4 |
Hb_001006_240 |
0.2974822048 |
- |
- |
- |
| 5 |
Hb_001545_210 |
0.3134602764 |
- |
- |
hypothetical protein JCGZ_11132 [Jatropha curcas] |
| 6 |
Hb_025311_020 |
0.3249494835 |
- |
- |
PREDICTED: uncharacterized protein LOC105039527 [Elaeis guineensis] |
| 7 |
Hb_001943_020 |
0.334678464 |
- |
- |
PREDICTED: (S)-coclaurine N-methyltransferase-like, partial [Populus euphratica] |
| 8 |
Hb_000165_150 |
0.3352792699 |
- |
- |
PREDICTED: pistil-specific extensin-like protein [Jatropha curcas] |
| 9 |
Hb_089419_010 |
0.3360885043 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 10 |
Hb_000190_240 |
0.338296791 |
- |
- |
unnamed protein product [Coffea canephora] |
| 11 |
Hb_001304_120 |
0.3408622761 |
transcription factor |
TF Family: WRKY |
WRKY protein [Hevea brasiliensis] |
| 12 |
Hb_000422_120 |
0.3420919867 |
- |
- |
PREDICTED: 1-phosphatidylinositol-3-phosphate 5-kinase FAB1B [Jatropha curcas] |
| 13 |
Hb_143874_010 |
0.3432450455 |
- |
- |
PREDICTED: uncharacterized protein LOC105794100 [Gossypium raimondii] |
| 14 |
Hb_000276_050 |
0.3449793931 |
- |
- |
- |
| 15 |
Hb_003688_160 |
0.3533281037 |
- |
- |
Presenilin-like family protein [Populus trichocarpa] |
| 16 |
Hb_006280_010 |
0.3538141386 |
transcription factor |
TF Family: ERF |
PREDICTED: AP2-like ethylene-responsive transcription factor TOE3 isoform X2 [Jatropha curcas] |
| 17 |
Hb_177179_010 |
0.3559072171 |
- |
- |
PREDICTED: putative 4-hydroxy-4-methyl-2-oxoglutarate aldolase 2 [Populus euphratica] |
| 18 |
Hb_006911_050 |
0.3597183268 |
- |
- |
PREDICTED: uncharacterized protein LOC105628750 [Jatropha curcas] |
| 19 |
Hb_001242_090 |
0.3631501882 |
- |
- |
40S ribosomal protein S18 [Colletotrichum fioriniae PJ7] |
| 20 |
Hb_003881_050 |
0.3642698589 |
- |
- |
PREDICTED: uncharacterized protein LOC104213140 [Nicotiana sylvestris] |