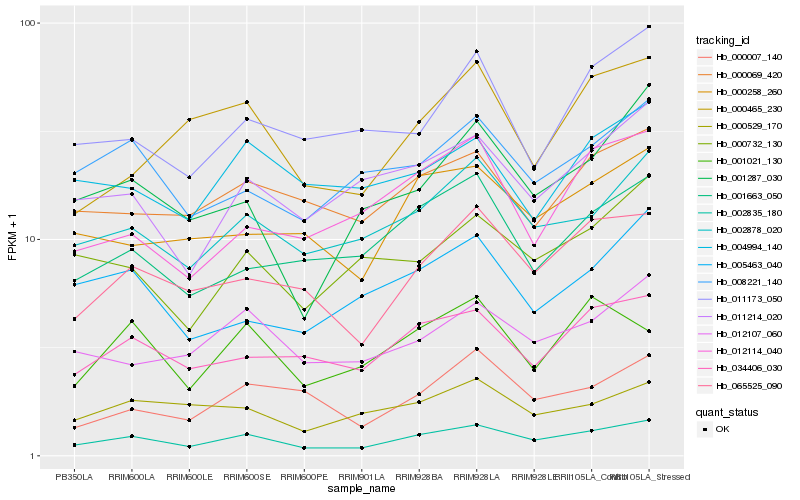
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002835_180 |
0.0 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 2 |
Hb_065525_090 |
0.096760262 |
- |
- |
PREDICTED: CASP-like protein 4C1 [Vitis vinifera] |
| 3 |
Hb_002878_020 |
0.0984744842 |
- |
- |
mitochondrial carrier protein, putative [Ricinus communis] |
| 4 |
Hb_034406_030 |
0.1059676002 |
- |
- |
PREDICTED: dihydroorotate dehydrogenase (quinone), mitochondrial [Musa acuminata subsp. malaccensis] |
| 5 |
Hb_012107_060 |
0.1130235072 |
- |
- |
hypothetical protein JCGZ_17178 [Jatropha curcas] |
| 6 |
Hb_011214_020 |
0.1147158256 |
- |
- |
PREDICTED: RNA-binding protein 5 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000258_260 |
0.1174913434 |
- |
- |
PREDICTED: protein HGH1 homolog [Jatropha curcas] |
| 8 |
Hb_001287_030 |
0.119586243 |
- |
- |
30S ribosomal protein S31, mitochondrial precursor, putative [Ricinus communis] |
| 9 |
Hb_000069_420 |
0.1195863424 |
- |
- |
Beclin-1, putative [Ricinus communis] |
| 10 |
Hb_008221_140 |
0.120593265 |
- |
- |
PREDICTED: uncharacterized protein LOC105644226 [Jatropha curcas] |
| 11 |
Hb_001663_050 |
0.1215377806 |
- |
- |
PREDICTED: GPN-loop GTPase 3 [Jatropha curcas] |
| 12 |
Hb_000732_130 |
0.1221877379 |
transcription factor |
TF Family: HSF |
hypothetical protein JCGZ_17108 [Jatropha curcas] |
| 13 |
Hb_000465_230 |
0.1256407725 |
- |
- |
PREDICTED: transcriptional regulator ATRX homolog [Jatropha curcas] |
| 14 |
Hb_000529_170 |
0.1256816508 |
- |
- |
hypothetical protein JCGZ_14691 [Jatropha curcas] |
| 15 |
Hb_001021_130 |
0.1266434254 |
- |
- |
PREDICTED: uncharacterized protein LOC105642479 [Jatropha curcas] |
| 16 |
Hb_000007_140 |
0.1280048504 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570 [Jatropha curcas] |
| 17 |
Hb_005463_040 |
0.1293826249 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 18 |
Hb_011173_050 |
0.1296378508 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 19 |
Hb_012114_040 |
0.130320363 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_004994_140 |
0.1303234355 |
- |
- |
PREDICTED: digalactosyldiacylglycerol synthase 2, chloroplastic [Jatropha curcas] |