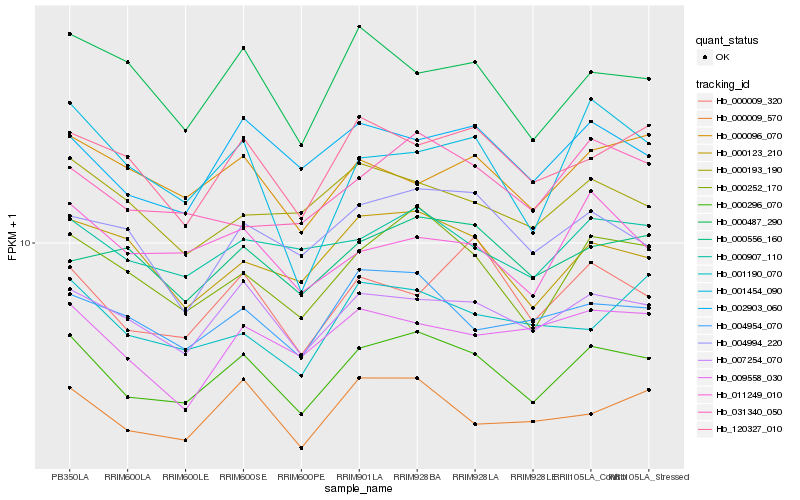
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000296_070 |
0.0 |
- |
- |
hypothetical protein B456_003G048700 [Gossypium raimondii] |
| 2 |
Hb_000252_170 |
0.0682599963 |
- |
- |
PREDICTED: uncharacterized protein LOC105644650 isoform X2 [Jatropha curcas] |
| 3 |
Hb_007254_070 |
0.0821446604 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HERC2-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_002903_060 |
0.0961877147 |
- |
- |
PREDICTED: uncharacterized protein LOC105649505 [Jatropha curcas] |
| 5 |
Hb_000123_210 |
0.0968000851 |
- |
- |
PREDICTED: uncharacterized protein LOC105641576 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000907_110 |
0.0971705998 |
- |
- |
dihydroxyacetone kinase family protein [Populus trichocarpa] |
| 7 |
Hb_120327_010 |
0.0998985292 |
- |
- |
PREDICTED: uncharacterized protein LOC105650959 [Jatropha curcas] |
| 8 |
Hb_004954_070 |
0.1010949341 |
- |
- |
PREDICTED: uncharacterized protein LOC105645187 [Jatropha curcas] |
| 9 |
Hb_000009_320 |
0.1032914966 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66631 [Jatropha curcas] |
| 10 |
Hb_004994_220 |
0.1046197661 |
- |
- |
PREDICTED: K(+) efflux antiporter 2, chloroplastic-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_031340_050 |
0.1048877528 |
- |
- |
PREDICTED: tropinone reductase 1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000009_570 |
0.106366737 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Populus euphratica] |
| 13 |
Hb_000487_290 |
0.106830485 |
- |
- |
ribonucleic acid binding protein S1, putative [Ricinus communis] |
| 14 |
Hb_001454_090 |
0.1069633711 |
- |
- |
PREDICTED: alanyl-tRNA editing protein Aarsd1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000556_160 |
0.1080736108 |
- |
- |
PREDICTED: ankyrin repeat and zinc finger domain-containing protein 1 [Jatropha curcas] |
| 16 |
Hb_011249_010 |
0.1104422461 |
- |
- |
PREDICTED: N-alpha-acetyltransferase 35, NatC auxiliary subunit isoform X1 [Jatropha curcas] |
| 17 |
Hb_000193_190 |
0.110787422 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 16 [Jatropha curcas] |
| 18 |
Hb_000096_070 |
0.111935361 |
- |
- |
PREDICTED: ruvB-like 2 [Jatropha curcas] |
| 19 |
Hb_001190_070 |
0.1120194282 |
- |
- |
PREDICTED: mitochondrial ubiquitin ligase activator of nfkb 1-A [Jatropha curcas] |
| 20 |
Hb_009558_030 |
0.1122906559 |
- |
- |
myb3r3, putative [Ricinus communis] |