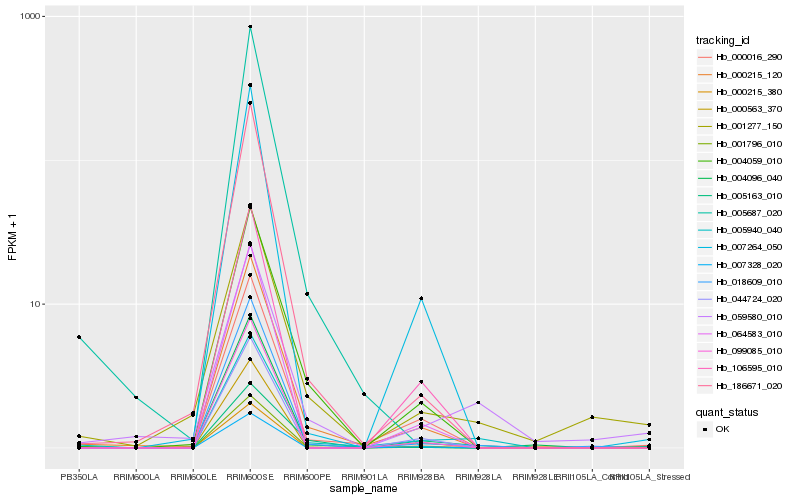
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005940_040 |
0.0 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 2 |
Hb_000215_120 |
0.1385399247 |
- |
- |
PREDICTED: putative ABC transporter B family member 8 [Jatropha curcas] |
| 3 |
Hb_000016_290 |
0.1425273487 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 4 |
Hb_059580_010 |
0.1442205968 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 13, plasma membrane-type [Jatropha curcas] |
| 5 |
Hb_018609_010 |
0.1445252986 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004059_010 |
0.1546245334 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 7 |
Hb_001796_010 |
0.1549241924 |
- |
- |
purine transporter, putative [Ricinus communis] |
| 8 |
Hb_005163_010 |
0.1582623005 |
- |
- |
PREDICTED: probable terpene synthase 11 [Jatropha curcas] |
| 9 |
Hb_001277_150 |
0.1598293249 |
- |
- |
hypothetical protein VITISV_043492 [Vitis vinifera] |
| 10 |
Hb_186671_020 |
0.1644488526 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_005687_020 |
0.1667860545 |
- |
- |
P66 protein [Hevea brasiliensis] |
| 12 |
Hb_007264_050 |
0.1670609799 |
- |
- |
- |
| 13 |
Hb_000563_370 |
0.1710403209 |
- |
- |
hypothetical protein JCGZ_18919 [Jatropha curcas] |
| 14 |
Hb_007328_020 |
0.1710890022 |
- |
- |
- |
| 15 |
Hb_000215_380 |
0.1711805294 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 16 |
Hb_064583_010 |
0.1715343988 |
- |
- |
PREDICTED: germin-like protein subfamily 1 member 7 [Vitis vinifera] |
| 17 |
Hb_044724_020 |
0.1723144817 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Fragaria vesca subsp. vesca] |
| 18 |
Hb_099085_010 |
0.1736428624 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 19 |
Hb_106595_010 |
0.1742633321 |
- |
- |
hypothetical protein B456_003G065300 [Gossypium raimondii] |
| 20 |
Hb_004096_040 |
0.1754031166 |
- |
- |
cytochrome P450, putative [Ricinus communis] |