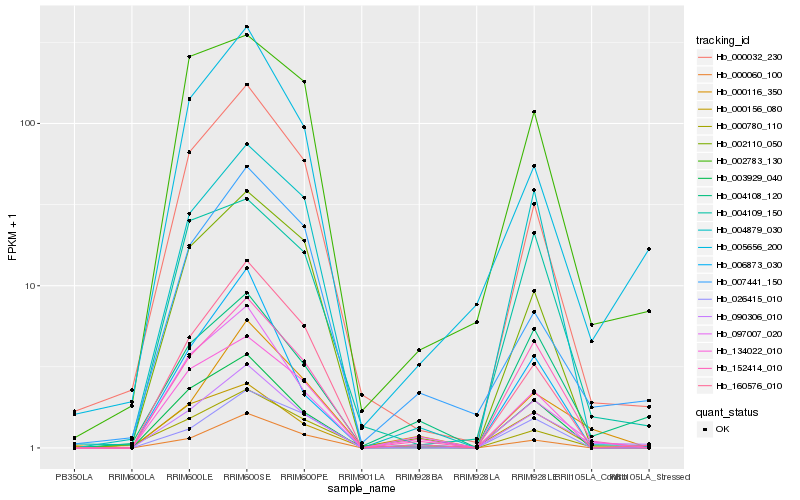
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_090306_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0007s02300g [Populus trichocarpa] |
| 2 |
Hb_152414_010 |
0.1306252745 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein PRUPE_ppa024232mg [Prunus persica] |
| 3 |
Hb_000032_230 |
0.136656726 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_20463 [Jatropha curcas] |
| 4 |
Hb_160576_010 |
0.1409104127 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 5 |
Hb_003929_040 |
0.1415384031 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_005656_200 |
0.1451295392 |
- |
- |
Cation transport protein chaC, putative [Ricinus communis] |
| 7 |
Hb_004879_030 |
0.1600369706 |
- |
- |
PREDICTED: uncharacterized protein LOC105124685 [Populus euphratica] |
| 8 |
Hb_000156_080 |
0.1610892766 |
- |
- |
Potassium channel KAT2, putative [Ricinus communis] |
| 9 |
Hb_000060_100 |
0.1620378534 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002110_050 |
0.1670689158 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_097007_020 |
0.1685688065 |
- |
- |
PREDICTED: nudix hydrolase 12, mitochondrial-like [Populus euphratica] |
| 12 |
Hb_004108_120 |
0.1694315728 |
- |
- |
PREDICTED: EID1-like F-box protein 3 [Jatropha curcas] |
| 13 |
Hb_006873_030 |
0.1699814837 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 14 |
Hb_000116_350 |
0.1721063573 |
- |
- |
PREDICTED: uncharacterized protein LOC105628582 [Jatropha curcas] |
| 15 |
Hb_000780_110 |
0.1726910412 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 16 |
Hb_134022_010 |
0.1728777346 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 17 |
Hb_004109_150 |
0.1746997516 |
- |
- |
glutathione-S-transferase tau 1 [Hevea brasiliensis] |
| 18 |
Hb_026415_010 |
0.1749571433 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002783_130 |
0.175043931 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_007441_150 |
0.179696473 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |