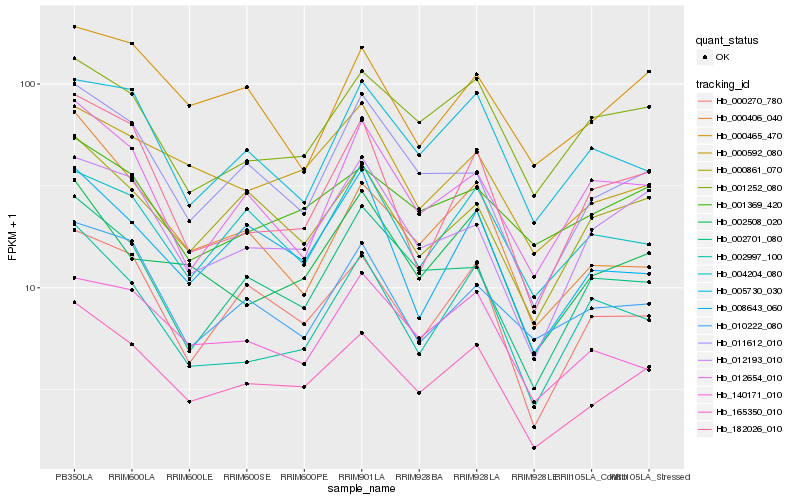
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_165350_010 |
0.0 |
- |
- |
unnamed protein product [Triticum aestivum] |
| 2 |
Hb_000270_780 |
0.0878975764 |
- |
- |
PREDICTED: probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 9 [Jatropha curcas] |
| 3 |
Hb_000861_070 |
0.0926649782 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_011612_010 |
0.0976034498 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 5 |
Hb_002508_020 |
0.0994458203 |
- |
- |
PREDICTED: SNF1-related protein kinase regulatory subunit beta-1 [Jatropha curcas] |
| 6 |
Hb_000592_080 |
0.101935293 |
- |
- |
PREDICTED: thymidylate kinase [Jatropha curcas] |
| 7 |
Hb_012193_010 |
0.103233376 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 8 |
Hb_002701_080 |
0.1069882588 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At3g02290-like [Jatropha curcas] |
| 9 |
Hb_001252_080 |
0.1082394367 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit G-like [Jatropha curcas] |
| 10 |
Hb_008643_060 |
0.1088213128 |
- |
- |
unnamed protein product [Coffea canephora] |
| 11 |
Hb_001369_420 |
0.1105012866 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIN2 [Jatropha curcas] |
| 12 |
Hb_010222_080 |
0.1115916379 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g06000 [Jatropha curcas] |
| 13 |
Hb_000465_470 |
0.1117402242 |
- |
- |
unknown [Picea sitchensis] |
| 14 |
Hb_140171_010 |
0.112099222 |
- |
- |
PREDICTED: magnesium transporter MRS2-3 [Jatropha curcas] |
| 15 |
Hb_182026_010 |
0.1128851582 |
- |
- |
hypothetical protein CICLE_v10003487mg, partial [Citrus clementina] |
| 16 |
Hb_004204_080 |
0.1132634966 |
- |
- |
diacylglycerol kinase, theta, putative [Ricinus communis] |
| 17 |
Hb_012654_010 |
0.1133292557 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105629849 [Jatropha curcas] |
| 18 |
Hb_002997_100 |
0.1145756296 |
- |
- |
hypothetical protein JCGZ_19555 [Jatropha curcas] |
| 19 |
Hb_000406_040 |
0.1151839396 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: FRIGIDA-like protein 5 [Jatropha curcas] |
| 20 |
Hb_005730_030 |
0.115277954 |
- |
- |
eukaryotic translation initiation factor 3 subunit, putative [Ricinus communis] |