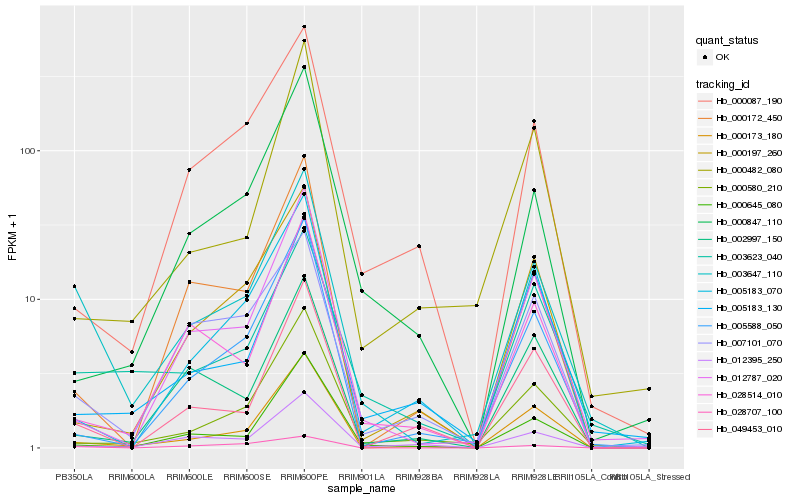
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003647_110 |
0.0 |
- |
- |
PREDICTED: 18.1 kDa class I heat shock protein-like [Populus euphratica] |
| 2 |
Hb_003623_040 |
0.179377107 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 3 |
Hb_000173_180 |
0.186180364 |
- |
- |
PREDICTED: adenylate isopentenyltransferase 5, chloroplastic [Jatropha curcas] |
| 4 |
Hb_028707_100 |
0.1958444613 |
- |
- |
hypothetical protein RCOM_1687010 [Ricinus communis] |
| 5 |
Hb_012787_020 |
0.2004931744 |
- |
- |
PREDICTED: thioredoxin H7-like [Jatropha curcas] |
| 6 |
Hb_000580_210 |
0.2047945731 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 15 [Jatropha curcas] |
| 7 |
Hb_007101_070 |
0.2065074619 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1 [Jatropha curcas] |
| 8 |
Hb_000645_080 |
0.2121036016 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 9 |
Hb_049453_010 |
0.2128609505 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_005588_050 |
0.2141169988 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_028514_010 |
0.2141396657 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A9 [Jatropha curcas] |
| 12 |
Hb_005183_130 |
0.2209218796 |
- |
- |
glycogenin, putative [Ricinus communis] |
| 13 |
Hb_000172_450 |
0.2214742527 |
- |
- |
Uncharacterized protein TCM_001203 [Theobroma cacao] |
| 14 |
Hb_000087_190 |
0.2237069933 |
- |
- |
hypothetical protein CICLE_v10033278mg [Citrus clementina] |
| 15 |
Hb_012395_250 |
0.2242129374 |
- |
- |
hypothetical protein RCOM_1217340 [Ricinus communis] |
| 16 |
Hb_000847_110 |
0.2263321339 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000197_260 |
0.2265663096 |
- |
- |
hypothetical protein PRUPE_ppa025448mg [Prunus persica] |
| 18 |
Hb_005183_070 |
0.227547482 |
- |
- |
PREDICTED: calmodulin-like protein 5 [Solanum lycopersicum] |
| 19 |
Hb_000482_080 |
0.2282924988 |
- |
- |
PREDICTED: uncharacterized protein LOC105638144 [Jatropha curcas] |
| 20 |
Hb_002997_150 |
0.2288468978 |
- |
- |
hypothetical protein RCOM_1406750 [Ricinus communis] |