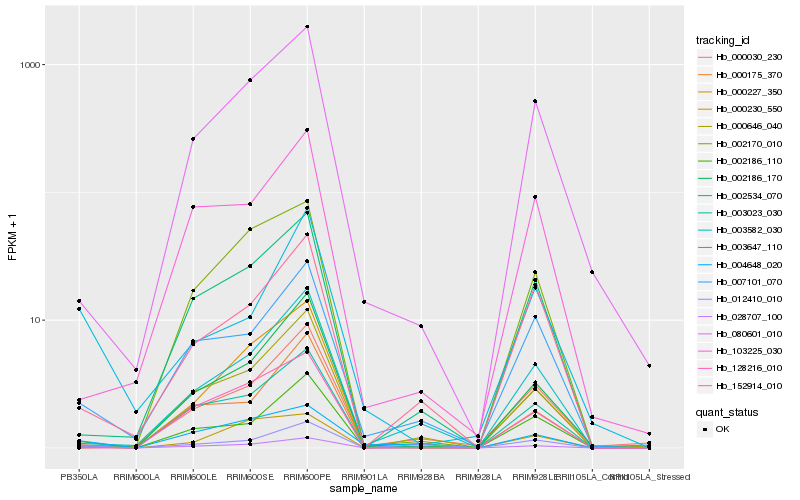
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028707_100 |
0.0 |
- |
- |
hypothetical protein RCOM_1687010 [Ricinus communis] |
| 2 |
Hb_002186_170 |
0.1624822343 |
- |
- |
hypothetical protein POPTR_0067s00200g [Populus trichocarpa] |
| 3 |
Hb_003023_030 |
0.1655168064 |
- |
- |
PREDICTED: MATE efflux family protein 5-like isoform X1 [Citrus sinensis] |
| 4 |
Hb_007101_070 |
0.1697529176 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1 [Jatropha curcas] |
| 5 |
Hb_080601_010 |
0.1739473998 |
- |
- |
metallothionein 3-like protein [Hevea brasiliensis] |
| 6 |
Hb_002186_110 |
0.1775788557 |
- |
- |
hypothetical protein CICLE_v10024045mg, partial [Citrus clementina] |
| 7 |
Hb_002534_070 |
0.1779872372 |
- |
- |
PREDICTED: uncharacterized protein LOC105640545 [Jatropha curcas] |
| 8 |
Hb_000175_370 |
0.182659415 |
- |
- |
hypothetical protein EUGRSUZ_F03862 [Eucalyptus grandis] |
| 9 |
Hb_000227_350 |
0.1839429735 |
- |
- |
hypothetical protein CICLE_v10006537mg [Citrus clementina] |
| 10 |
Hb_004648_020 |
0.1851577507 |
- |
- |
PREDICTED: uncharacterized protein LOC105643062 [Jatropha curcas] |
| 11 |
Hb_128216_010 |
0.1884779211 |
- |
- |
PREDICTED: UDP-glycosyltransferase 79B6-like [Jatropha curcas] |
| 12 |
Hb_152914_010 |
0.1907588828 |
- |
- |
PREDICTED: polcalcin Syr v 3-like [Nicotiana sylvestris] |
| 13 |
Hb_003582_030 |
0.1919373996 |
- |
- |
PREDICTED: serine/threonine-protein kinase BRI1-like 2 [Jatropha curcas] |
| 14 |
Hb_012410_010 |
0.1936552157 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 15 |
Hb_000646_040 |
0.1942010334 |
- |
- |
PREDICTED: thiamine-repressible mitochondrial transport protein THI74-like [Jatropha curcas] |
| 16 |
Hb_000230_550 |
0.1944564356 |
- |
- |
PREDICTED: adenylate isopentenyltransferase 5, chloroplastic [Jatropha curcas] |
| 17 |
Hb_003647_110 |
0.1958444613 |
- |
- |
PREDICTED: 18.1 kDa class I heat shock protein-like [Populus euphratica] |
| 18 |
Hb_000030_230 |
0.196196747 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 19 |
Hb_002170_010 |
0.1965118134 |
- |
- |
hypothetical protein JCGZ_25046 [Jatropha curcas] |
| 20 |
Hb_103225_030 |
0.1968775513 |
transcription factor |
TF Family: Orphans |
DNA binding protein, putative [Ricinus communis] |