| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
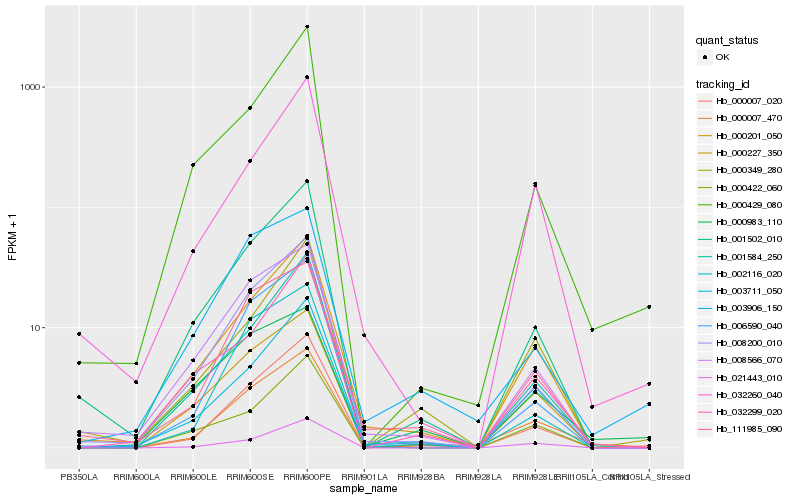
Hb_000007_470 |
0.0 |
- |
- |
PREDICTED: phosphate transporter PHO1 homolog 1 [Jatropha curcas] |
| 2 |
Hb_000007_020 |
0.1182118829 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 3 |
Hb_000201_050 |
0.1272428495 |
- |
- |
- |
| 4 |
Hb_000227_350 |
0.133736456 |
- |
- |
hypothetical protein CICLE_v10006537mg [Citrus clementina] |
| 5 |
Hb_021443_010 |
0.1381840896 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 1 [Jatropha curcas] |
| 6 |
Hb_003711_050 |
0.1402224402 |
- |
- |
Isoflavone reductase, putative [Ricinus communis] |
| 7 |
Hb_111985_090 |
0.1407231736 |
- |
- |
hypothetical protein RCOM_1595950 [Ricinus communis] |
| 8 |
Hb_001502_010 |
0.1415304995 |
- |
- |
hypothetical protein JCGZ_04288 [Jatropha curcas] |
| 9 |
Hb_008200_010 |
0.1421533322 |
- |
- |
PREDICTED: ABC transporter G family member 14-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_001584_250 |
0.1435848038 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 11 |
Hb_006590_040 |
0.1458492713 |
- |
- |
amino acid permease, putative [Ricinus communis] |
| 12 |
Hb_003906_150 |
0.1488753403 |
- |
- |
peptidoglycan-binding LysM domain-containing family protein, partial [Populus trichocarpa] |
| 13 |
Hb_008566_070 |
0.1491479671 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_032260_040 |
0.1528271841 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000349_280 |
0.1528447768 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002116_020 |
0.1546313222 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 35 [Jatropha curcas] |
| 17 |
Hb_000422_060 |
0.1552518926 |
transcription factor |
TF Family: OFP |
hypothetical protein RCOM_1282480 [Ricinus communis] |
| 18 |
Hb_032299_020 |
0.1563396755 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 19 |
Hb_000429_080 |
0.157787046 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000983_110 |
0.1597100565 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |