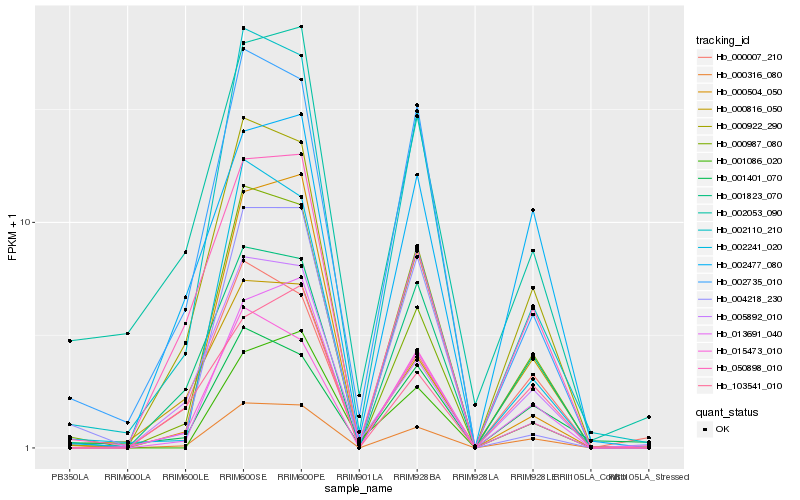
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000316_080 |
0.0 |
- |
- |
ENTH/VHS family protein [Theobroma cacao] |
| 2 |
Hb_000987_080 |
0.0962279111 |
- |
- |
light-inducible protein atls1, putative [Ricinus communis] |
| 3 |
Hb_013691_040 |
0.1197244117 |
- |
- |
hypothetical protein B456_001G045900 [Gossypium raimondii] |
| 4 |
Hb_000922_290 |
0.120740608 |
- |
- |
PREDICTED: probable boron transporter 2 [Jatropha curcas] |
| 5 |
Hb_050898_010 |
0.123359438 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 6 |
Hb_001823_070 |
0.1289467432 |
- |
- |
PREDICTED: uncharacterized protein LOC105637371 [Jatropha curcas] |
| 7 |
Hb_002110_210 |
0.1382193357 |
- |
- |
purine transporter, putative [Ricinus communis] |
| 8 |
Hb_002053_090 |
0.1396119196 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002241_020 |
0.1404115226 |
- |
- |
PREDICTED: salicylic acid-binding protein 2-like [Jatropha curcas] |
| 10 |
Hb_000007_210 |
0.1456494026 |
- |
- |
hypothetical protein POPTR_0010s14360g [Populus trichocarpa] |
| 11 |
Hb_005892_010 |
0.1457185612 |
- |
- |
PREDICTED: uncharacterized protein LOC105632168 [Jatropha curcas] |
| 12 |
Hb_002735_010 |
0.1477823348 |
- |
- |
PREDICTED: CLAVATA3/ESR (CLE)-related protein 44 [Prunus mume] |
| 13 |
Hb_000816_050 |
0.1569346089 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 14 |
Hb_015473_010 |
0.1577663627 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 15 |
Hb_002477_080 |
0.1610897675 |
- |
- |
hypothetical protein RCOM_0653450 [Ricinus communis] |
| 16 |
Hb_103541_010 |
0.1684395961 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X2 [Jatropha curcas] |
| 17 |
Hb_004218_230 |
0.1692781308 |
- |
- |
Peptide transporter, putative [Ricinus communis] |
| 18 |
Hb_001401_070 |
0.1696823187 |
- |
- |
glycogenin, putative [Ricinus communis] |
| 19 |
Hb_001086_020 |
0.1699748393 |
- |
- |
hypothetical protein POPTR_0013s01730g [Populus trichocarpa] |
| 20 |
Hb_000504_050 |
0.1703328925 |
transcription factor |
TF Family: LOB |
LOB domain protein 25 [Populus trichocarpa] |