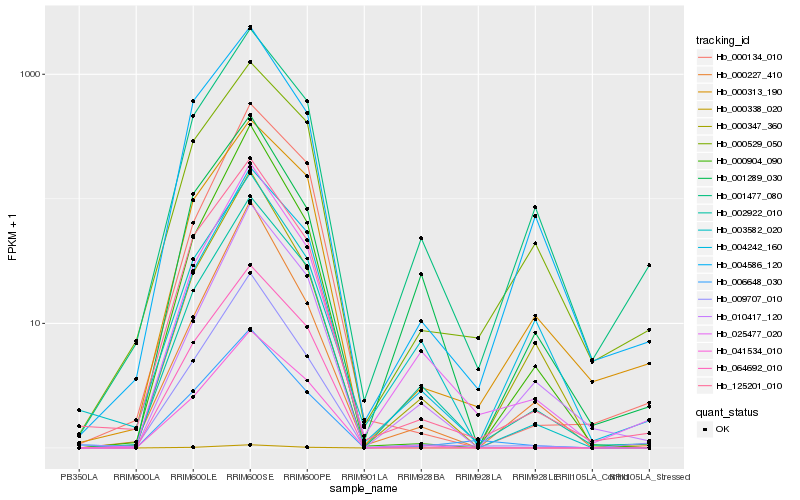
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002922_010 |
0.0 |
- |
- |
heavy-metal-associated domain-containing family protein [Populus trichocarpa] |
| 2 |
Hb_025477_020 |
0.0617868242 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF027 [Jatropha curcas] |
| 3 |
Hb_010417_120 |
0.0858582535 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001477_080 |
0.0890512703 |
- |
- |
hypothetical protein POPTR_0017s04860g [Populus trichocarpa] |
| 5 |
Hb_041534_010 |
0.0892014052 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF017 [Jatropha curcas] |
| 6 |
Hb_125201_010 |
0.0967578519 |
- |
- |
hypothetical protein POPTR_0004s03690g [Populus trichocarpa] |
| 7 |
Hb_003582_020 |
0.0968839159 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Eucalyptus grandis] |
| 8 |
Hb_000313_190 |
0.1006257995 |
- |
- |
PREDICTED: uncharacterized protein LOC105636896 [Jatropha curcas] |
| 9 |
Hb_004242_160 |
0.1035777127 |
transcription factor |
TF Family: GRAS |
PREDICTED: chitin-inducible gibberellin-responsive protein 1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_009707_010 |
0.1049287234 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: exocyst complex component EXO70B1-like [Jatropha curcas] |
| 11 |
Hb_000529_050 |
0.1055100656 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 12 |
Hb_001289_030 |
0.1078858134 |
- |
- |
syntaxin, plant, putative [Ricinus communis] |
| 13 |
Hb_000347_360 |
0.1083263669 |
- |
- |
PREDICTED: linoleate 13S-lipoxygenase 3-1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_004586_120 |
0.1122518058 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 15 |
Hb_006648_030 |
0.1124497859 |
- |
- |
Indole-3-acetic acid-amido synthetase GH3.3, putative [Ricinus communis] |
| 16 |
Hb_064692_010 |
0.1124876577 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 17 |
Hb_000338_020 |
0.1177835161 |
- |
- |
PREDICTED: tryptophan synthase beta chain 1 [Jatropha curcas] |
| 18 |
Hb_000904_090 |
0.1177908147 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF109-like [Jatropha curcas] |
| 19 |
Hb_000134_010 |
0.1182435783 |
- |
- |
- |
| 20 |
Hb_000227_410 |
0.1185768954 |
- |
- |
PREDICTED: uncharacterized protein LOC105631970 [Jatropha curcas] |