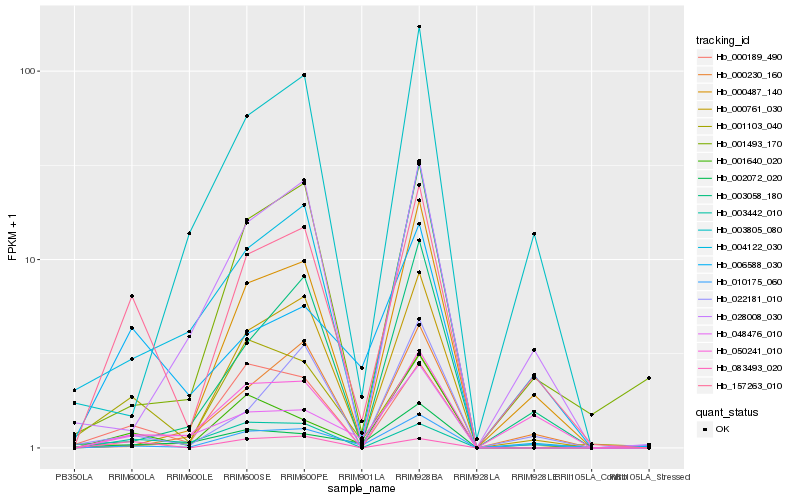
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_157263_010 |
0.0 |
- |
- |
PREDICTED: monomethylxanthine methyltransferase 2-like isoform X4 [Jatropha curcas] |
| 2 |
Hb_010175_060 |
0.166683576 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_050241_010 |
0.1995651311 |
- |
- |
PREDICTED: uncharacterized protein LOC105129233 isoform X1 [Populus euphratica] |
| 4 |
Hb_006588_030 |
0.210169944 |
- |
- |
PREDICTED: cytochrome P450 CYP72A219-like [Jatropha curcas] |
| 5 |
Hb_000189_490 |
0.2165236631 |
- |
- |
Sulfate/thiosulfate import ATP-binding protein cysA [Theobroma cacao] |
| 6 |
Hb_001493_170 |
0.2212718116 |
- |
- |
PREDICTED: uncharacterized protein LOC105631328 [Jatropha curcas] |
| 7 |
Hb_004122_030 |
0.2213331369 |
transcription factor |
TF Family: HB |
PREDICTED: WUSCHEL-related homeobox 4-like [Populus euphratica] |
| 8 |
Hb_022181_010 |
0.2214667317 |
- |
- |
PREDICTED: peroxidase 19 [Jatropha curcas] |
| 9 |
Hb_001640_020 |
0.2257086942 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG1-like [Jatropha curcas] |
| 10 |
Hb_048476_010 |
0.2262126689 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 1-like isoform X1 [Citrus sinensis] |
| 11 |
Hb_083493_020 |
0.2264962462 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Vitis vinifera] |
| 12 |
Hb_000487_140 |
0.2291678418 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 13 |
Hb_003058_180 |
0.2306473195 |
transcription factor |
TF Family: HSF |
hypothetical protein POPTR_0001s28040g [Populus trichocarpa] |
| 14 |
Hb_000761_030 |
0.2315503106 |
transcription factor |
TF Family: ERF |
Transcriptional factor TINY, putative [Ricinus communis] |
| 15 |
Hb_000230_160 |
0.2371987256 |
- |
- |
PREDICTED: uncharacterized protein LOC105631858 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002072_020 |
0.2419110999 |
- |
- |
hypothetical protein JCGZ_16057 [Jatropha curcas] |
| 17 |
Hb_003805_080 |
0.2428637553 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 18 |
Hb_001103_040 |
0.2437386812 |
- |
- |
hypothetical protein POPTR_0013s11170g [Populus trichocarpa] |
| 19 |
Hb_028008_030 |
0.2440277545 |
- |
- |
probable xyloglucan endotransglucosylase/hydrolase protein 23 precursor [Malus domestica] |
| 20 |
Hb_003442_010 |
0.2448260479 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 4-like isoform X2 [Citrus sinensis] |