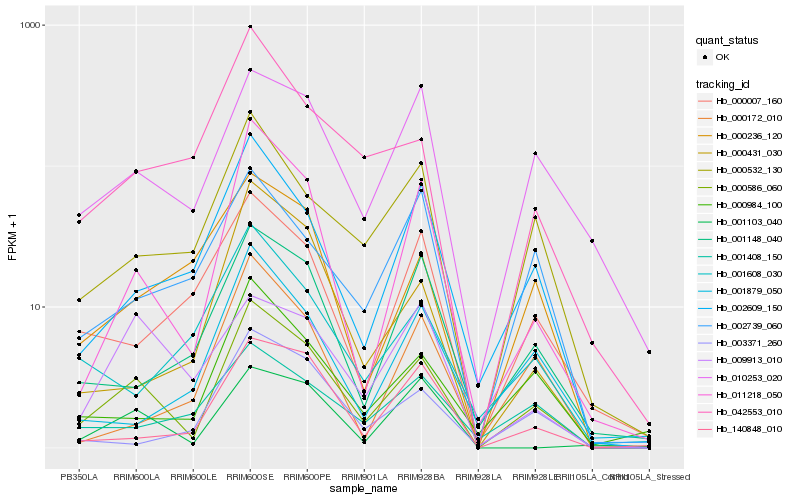
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000586_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670-like isoform X1 [Populus euphratica] |
| 2 |
Hb_011218_050 |
0.1676827071 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 3 |
Hb_000532_130 |
0.1908438465 |
- |
- |
hypothetical protein [Anabaena sp. PCC 7108] |
| 4 |
Hb_002609_150 |
0.1925031144 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000984_100 |
0.1978082192 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 6 |
Hb_010253_020 |
0.2009676377 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_001148_040 |
0.2097510299 |
- |
- |
hypothetical protein JCGZ_07831 [Jatropha curcas] |
| 8 |
Hb_000431_030 |
0.2103297355 |
transcription factor |
TF Family: C2H2 |
zinc finger family protein [Populus trichocarpa] |
| 9 |
Hb_042553_010 |
0.2128677844 |
- |
- |
PREDICTED: glutathione S-transferase T1-like [Jatropha curcas] |
| 10 |
Hb_140848_010 |
0.2135915578 |
- |
- |
hypothetical protein POPTR_0014s17480g [Populus trichocarpa] |
| 11 |
Hb_001103_040 |
0.2159240474 |
- |
- |
hypothetical protein POPTR_0013s11170g [Populus trichocarpa] |
| 12 |
Hb_000236_120 |
0.2161026332 |
- |
- |
- |
| 13 |
Hb_003371_260 |
0.2197117207 |
- |
- |
hypothetical protein EUGRSUZ_B03661 [Eucalyptus grandis] |
| 14 |
Hb_001408_150 |
0.2204655648 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000007_160 |
0.2220978166 |
- |
- |
hypothetical protein POPTR_0008s10970g [Populus trichocarpa] |
| 16 |
Hb_001879_050 |
0.2289492605 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 17 |
Hb_000172_010 |
0.2294961375 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670-like isoform X1 [Populus euphratica] |
| 18 |
Hb_001608_030 |
0.2309733713 |
- |
- |
Protein NSP-INTERACTING KINASE 1 [Glycine soja] |
| 19 |
Hb_002739_060 |
0.2312492224 |
- |
- |
- |
| 20 |
Hb_009913_010 |
0.2320363963 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |