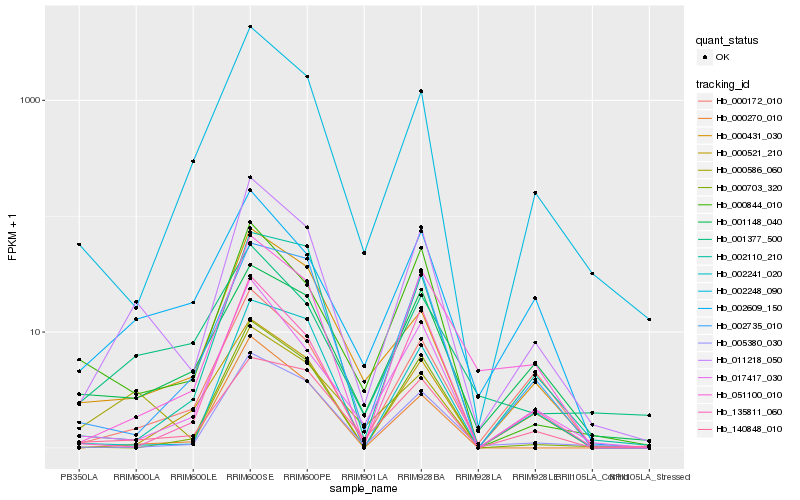
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011218_050 |
0.0 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 2 |
Hb_005380_030 |
0.1546520144 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase B120 isoform X2 [Jatropha curcas] |
| 3 |
Hb_017417_030 |
0.1626771138 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 4 |
Hb_002248_090 |
0.1669225094 |
- |
- |
metallothionein 3-like protein [Hevea brasiliensis] |
| 5 |
Hb_000586_060 |
0.1676827071 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670-like isoform X1 [Populus euphratica] |
| 6 |
Hb_051100_010 |
0.1686991779 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 7 |
Hb_002609_150 |
0.170152627 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_135811_060 |
0.1702016701 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 9 |
Hb_000172_010 |
0.1707203602 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670-like isoform X1 [Populus euphratica] |
| 10 |
Hb_000431_030 |
0.1715350686 |
transcription factor |
TF Family: C2H2 |
zinc finger family protein [Populus trichocarpa] |
| 11 |
Hb_000703_320 |
0.1740324325 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 12 |
Hb_140848_010 |
0.1743326258 |
- |
- |
hypothetical protein POPTR_0014s17480g [Populus trichocarpa] |
| 13 |
Hb_000270_010 |
0.1759784465 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001148_040 |
0.1804371124 |
- |
- |
hypothetical protein JCGZ_07831 [Jatropha curcas] |
| 15 |
Hb_000844_010 |
0.1823302076 |
- |
- |
PREDICTED: lysine histidine transporter-like 8 [Jatropha curcas] |
| 16 |
Hb_000521_210 |
0.1826994779 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 17 |
Hb_002241_020 |
0.1855952844 |
- |
- |
PREDICTED: salicylic acid-binding protein 2-like [Jatropha curcas] |
| 18 |
Hb_002735_010 |
0.1860402593 |
- |
- |
PREDICTED: CLAVATA3/ESR (CLE)-related protein 44 [Prunus mume] |
| 19 |
Hb_001377_500 |
0.1877655882 |
- |
- |
DOF AFFECTING GERMINATION 2 family protein [Populus trichocarpa] |
| 20 |
Hb_002110_210 |
0.1892615467 |
- |
- |
purine transporter, putative [Ricinus communis] |