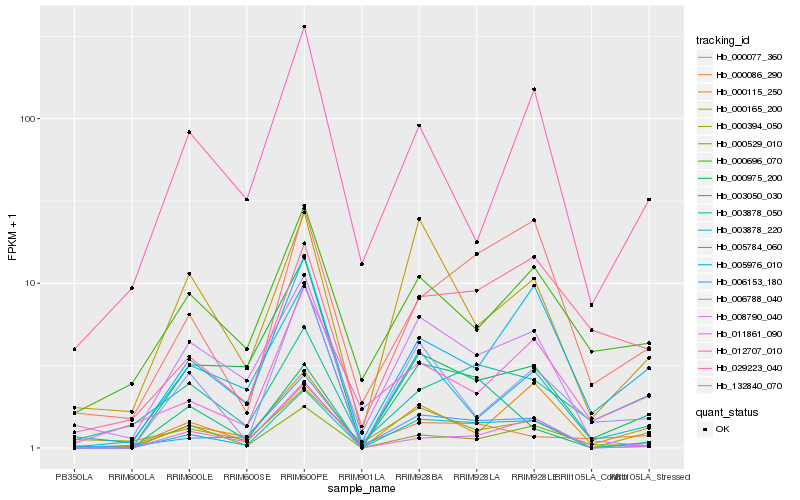
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003878_220 |
0.0 |
- |
- |
vacuolar protein sorting, putative [Ricinus communis] |
| 2 |
Hb_006153_180 |
0.1627212956 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_005976_010 |
0.2144887791 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_011861_090 |
0.2168074527 |
- |
- |
PREDICTED: probable sodium-coupled neutral amino acid transporter 6 [Jatropha curcas] |
| 5 |
Hb_008790_040 |
0.2175046222 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG8 [Jatropha curcas] |
| 6 |
Hb_000077_360 |
0.2197213538 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 7 |
Hb_000115_250 |
0.2222563951 |
- |
- |
PREDICTED: endonuclease 2 [Jatropha curcas] |
| 8 |
Hb_006788_040 |
0.2230303434 |
- |
- |
calmodulin, putative [Ricinus communis] |
| 9 |
Hb_000529_010 |
0.2247314806 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 10 |
Hb_000975_200 |
0.231123936 |
- |
- |
PREDICTED: uncharacterized protein LOC105628231 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000165_200 |
0.2348399888 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 12 |
Hb_003050_030 |
0.2467211426 |
- |
- |
Calcyclin-binding protein, putative [Ricinus communis] |
| 13 |
Hb_000394_050 |
0.2485039208 |
- |
- |
- |
| 14 |
Hb_005784_060 |
0.2507672747 |
- |
- |
PREDICTED: U-box domain-containing protein 28 [Jatropha curcas] |
| 15 |
Hb_012707_010 |
0.2526680617 |
- |
- |
PREDICTED: uncharacterized protein C594.04c [Jatropha curcas] |
| 16 |
Hb_003878_050 |
0.2605685115 |
- |
- |
PREDICTED: peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase A-like [Jatropha curcas] |
| 17 |
Hb_000086_290 |
0.2607265651 |
- |
- |
PREDICTED: TBC1 domain family member 15-like [Jatropha curcas] |
| 18 |
Hb_132840_070 |
0.2608118689 |
- |
- |
metal tolerance protein 1 [Populus trichocarpa x Populus deltoides] |
| 19 |
Hb_000696_070 |
0.2616906485 |
- |
- |
PREDICTED: aspartic proteinase-like protein 1 [Jatropha curcas] |
| 20 |
Hb_029223_040 |
0.2618738211 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |