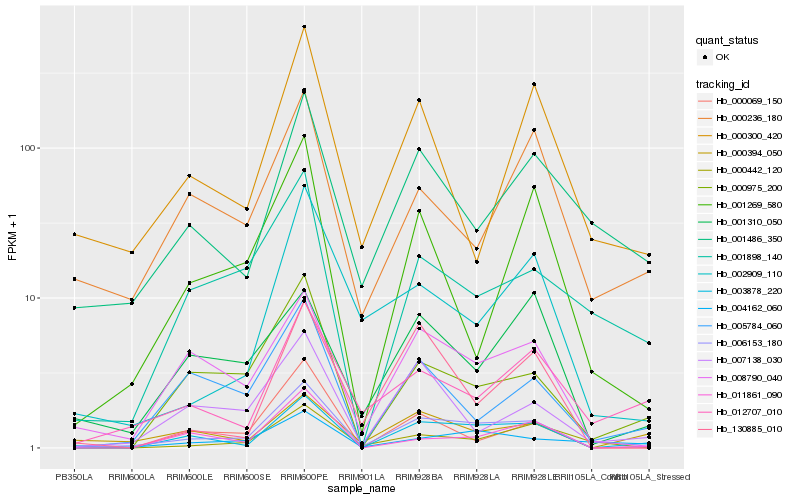
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006153_180 |
0.0 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.3 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003878_220 |
0.1627212956 |
- |
- |
vacuolar protein sorting, putative [Ricinus communis] |
| 3 |
Hb_008790_040 |
0.1750844853 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG8 [Jatropha curcas] |
| 4 |
Hb_000975_200 |
0.2056859456 |
- |
- |
PREDICTED: uncharacterized protein LOC105628231 isoform X2 [Jatropha curcas] |
| 5 |
Hb_011861_090 |
0.220505532 |
- |
- |
PREDICTED: probable sodium-coupled neutral amino acid transporter 6 [Jatropha curcas] |
| 6 |
Hb_000069_150 |
0.2215658718 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 7 |
Hb_002909_110 |
0.2240503117 |
- |
- |
PREDICTED: uncharacterized protein LOC105648153 [Jatropha curcas] |
| 8 |
Hb_005784_060 |
0.2261338348 |
- |
- |
PREDICTED: U-box domain-containing protein 28 [Jatropha curcas] |
| 9 |
Hb_001269_580 |
0.2312105515 |
- |
- |
PREDICTED: shikimate O-hydroxycinnamoyltransferase [Jatropha curcas] |
| 10 |
Hb_007138_030 |
0.2316396535 |
- |
- |
PREDICTED: putative disease resistance protein At4g11170 [Jatropha curcas] |
| 11 |
Hb_130885_010 |
0.231959544 |
- |
- |
PREDICTED: beta-glucosidase 13-like [Jatropha curcas] |
| 12 |
Hb_001898_140 |
0.2356470935 |
- |
- |
hypothetical protein AMTR_s00071p00184460 [Amborella trichopoda] |
| 13 |
Hb_000442_120 |
0.2368321879 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_012707_010 |
0.2384402427 |
- |
- |
PREDICTED: uncharacterized protein C594.04c [Jatropha curcas] |
| 15 |
Hb_004162_060 |
0.2394350267 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_001486_350 |
0.2394634927 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |
| 17 |
Hb_001310_050 |
0.2419522087 |
- |
- |
PREDICTED: formin-like protein 6 [Jatropha curcas] |
| 18 |
Hb_000394_050 |
0.2427358804 |
- |
- |
- |
| 19 |
Hb_000236_180 |
0.2442412447 |
- |
- |
serine hydroxymethyltransferase, putative [Ricinus communis] |
| 20 |
Hb_000300_420 |
0.2463650584 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |