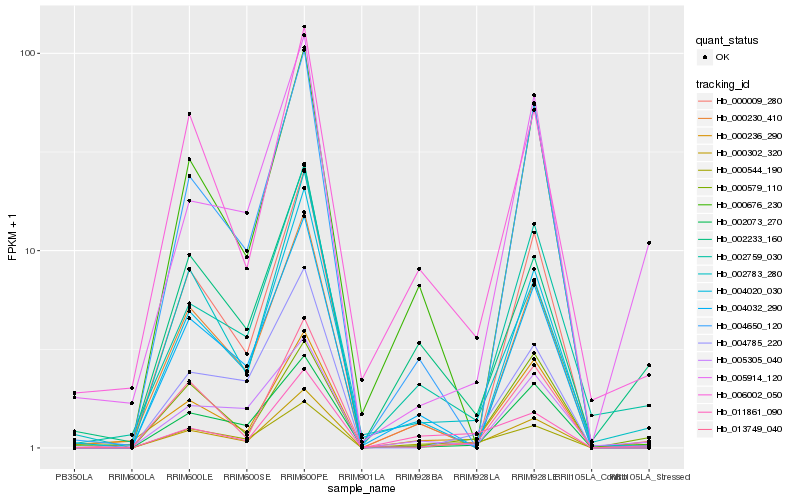
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000302_320 |
0.0 |
- |
- |
hypothetical protein JCGZ_26057 [Jatropha curcas] |
| 2 |
Hb_002759_030 |
0.1285381959 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1 [Jatropha curcas] |
| 3 |
Hb_000544_190 |
0.1360715181 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 4 |
Hb_000236_290 |
0.140380638 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 5 |
Hb_011861_090 |
0.1407979542 |
- |
- |
PREDICTED: probable sodium-coupled neutral amino acid transporter 6 [Jatropha curcas] |
| 6 |
Hb_002783_280 |
0.143033762 |
- |
- |
PREDICTED: WEB family protein At1g75720 [Jatropha curcas] |
| 7 |
Hb_000009_280 |
0.1467701665 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 8 |
Hb_006002_050 |
0.1469543214 |
- |
- |
PREDICTED: uncharacterized protein LOC105638083 [Jatropha curcas] |
| 9 |
Hb_005305_040 |
0.1476933804 |
transcription factor |
TF Family: C3H |
transcription factor, putative [Ricinus communis] |
| 10 |
Hb_000230_410 |
0.1483864458 |
- |
- |
PREDICTED: uncharacterized protein LOC105631891 [Jatropha curcas] |
| 11 |
Hb_000579_110 |
0.1488258804 |
- |
- |
PREDICTED: transmembrane protein 136 [Jatropha curcas] |
| 12 |
Hb_002073_270 |
0.1535703557 |
desease resistance |
Gene Name: TIR |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 13 |
Hb_013749_040 |
0.1576023383 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase PXL1 [Jatropha curcas] |
| 14 |
Hb_002233_160 |
0.1577320249 |
- |
- |
PREDICTED: uncharacterized protein LOC105633094 [Jatropha curcas] |
| 15 |
Hb_004032_290 |
0.1578887875 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 16 |
Hb_004650_120 |
0.1582264523 |
- |
- |
PREDICTED: auxin transporter-like protein 3 [Jatropha curcas] |
| 17 |
Hb_005914_120 |
0.1584566019 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 2B [Jatropha curcas] |
| 18 |
Hb_004785_220 |
0.158458913 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.2 [Populus euphratica] |
| 19 |
Hb_004020_030 |
0.1587620208 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 20 |
Hb_000676_230 |
0.1620886729 |
- |
- |
PREDICTED: beta-galactosidase 3 [Jatropha curcas] |