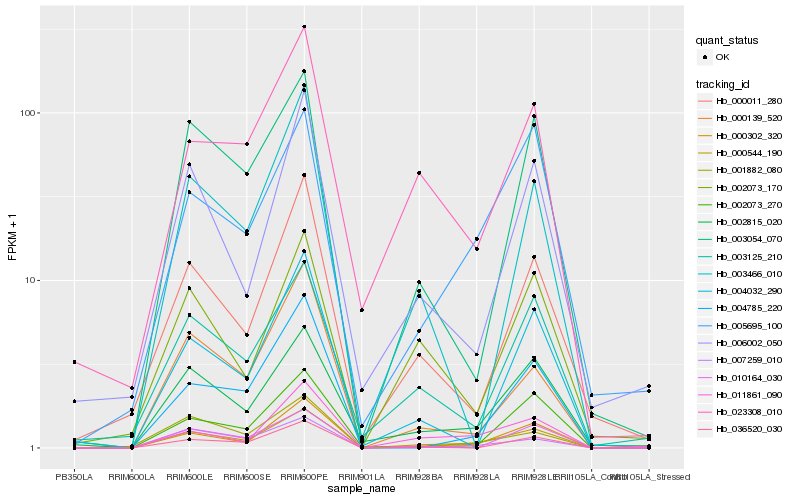
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000544_190 |
0.0 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 2 |
Hb_002815_020 |
0.1211371055 |
transcription factor |
TF Family: G2-like |
Homeodomain-like superfamily protein isoform 2 [Theobroma cacao] |
| 3 |
Hb_000302_320 |
0.1360715181 |
- |
- |
hypothetical protein JCGZ_26057 [Jatropha curcas] |
| 4 |
Hb_003054_070 |
0.1485609154 |
transcription factor |
TF Family: Orphans |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_003125_210 |
0.1542864799 |
- |
- |
Phospholipase C 4 precursor, putative [Ricinus communis] |
| 6 |
Hb_023308_010 |
0.1544764481 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 7 |
Hb_011861_090 |
0.1548040981 |
- |
- |
PREDICTED: probable sodium-coupled neutral amino acid transporter 6 [Jatropha curcas] |
| 8 |
Hb_005695_100 |
0.1566736119 |
- |
- |
PREDICTED: uncharacterized protein LOC105638839 [Jatropha curcas] |
| 9 |
Hb_000011_280 |
0.158350305 |
- |
- |
pectinesterase family protein [Populus trichocarpa] |
| 10 |
Hb_000139_520 |
0.1630984688 |
- |
- |
EF-hand calcium-binding domain-containing protein 4A [Theobroma cacao] |
| 11 |
Hb_001882_080 |
0.1637977344 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC103324347 [Prunus mume] |
| 12 |
Hb_010164_030 |
0.1653336296 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_004032_290 |
0.1654406106 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_006002_050 |
0.165576369 |
- |
- |
PREDICTED: uncharacterized protein LOC105638083 [Jatropha curcas] |
| 15 |
Hb_002073_170 |
0.167121006 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g27190 [Jatropha curcas] |
| 16 |
Hb_036520_030 |
0.1680249905 |
- |
- |
PREDICTED: uncharacterized protein LOC105638098 [Jatropha curcas] |
| 17 |
Hb_003466_010 |
0.1700737918 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 2.11-like [Jatropha curcas] |
| 18 |
Hb_002073_270 |
0.1709425533 |
desease resistance |
Gene Name: TIR |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 19 |
Hb_004785_220 |
0.1710586188 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.2 [Populus euphratica] |
| 20 |
Hb_007259_010 |
0.1714884868 |
- |
- |
hypothetical protein CISIN_1g0073972mg, partial [Citrus sinensis] |