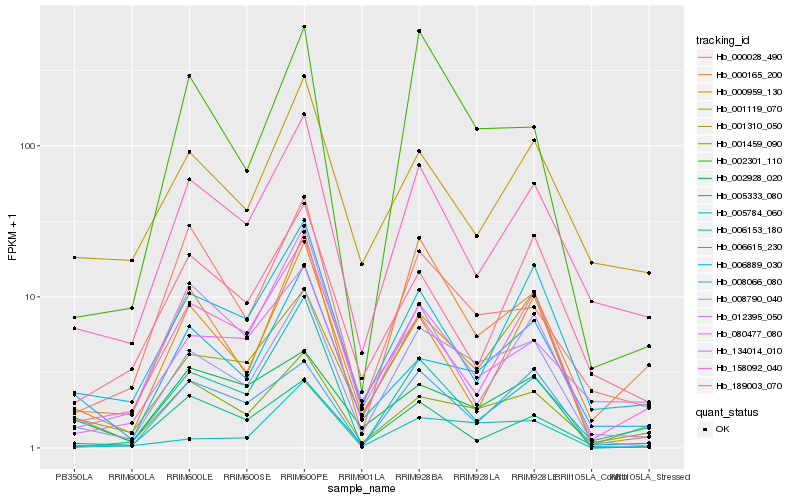
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008790_040 |
0.0 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG8 [Jatropha curcas] |
| 2 |
Hb_158092_040 |
0.1565979113 |
- |
- |
PREDICTED: uncharacterized protein LOC105647294 [Jatropha curcas] |
| 3 |
Hb_001310_050 |
0.1656696251 |
- |
- |
PREDICTED: formin-like protein 6 [Jatropha curcas] |
| 4 |
Hb_008066_080 |
0.1704650451 |
- |
- |
PREDICTED: GDSL esterase/lipase 1-like [Jatropha curcas] |
| 5 |
Hb_006889_030 |
0.1746438904 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |
| 6 |
Hb_006153_180 |
0.1750844853 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002301_110 |
0.1779872178 |
- |
- |
PREDICTED: protein E6-like [Populus euphratica] |
| 8 |
Hb_006615_230 |
0.1804906174 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 9 |
Hb_000165_200 |
0.1819583567 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 10 |
Hb_001459_090 |
0.1819852591 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_080477_080 |
0.1826032008 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 isoform X1 [Gossypium raimondii] |
| 12 |
Hb_000959_130 |
0.1876316935 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 13 |
Hb_001119_070 |
0.1891168676 |
- |
- |
methylenetetrahydrofolate reductase, putative [Ricinus communis] |
| 14 |
Hb_134014_010 |
0.1911821361 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 15 |
Hb_000028_490 |
0.194091505 |
- |
- |
calmodulin-like protein 6a [Populus trichocarpa] |
| 16 |
Hb_002928_020 |
0.1948093267 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 17 |
Hb_005333_080 |
0.1986089133 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005784_060 |
0.199634041 |
- |
- |
PREDICTED: U-box domain-containing protein 28 [Jatropha curcas] |
| 19 |
Hb_012395_050 |
0.2035998605 |
- |
- |
PREDICTED: uncharacterized protein LOC105632180 [Jatropha curcas] |
| 20 |
Hb_189003_070 |
0.2044462788 |
- |
- |
PREDICTED: peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase A-like [Jatropha curcas] |