| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
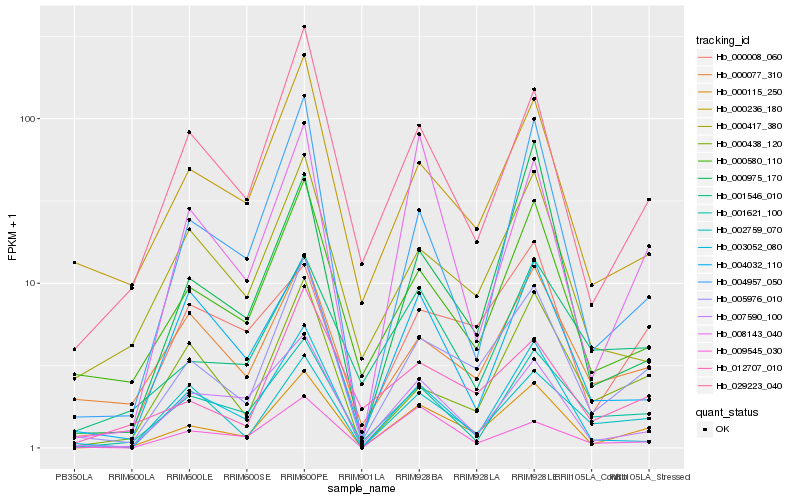
Hb_000115_250 |
0.0 |
- |
- |
PREDICTED: endonuclease 2 [Jatropha curcas] |
| 2 |
Hb_005976_010 |
0.1027099104 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_008143_040 |
0.1670709498 |
- |
- |
Alpha-expansin 4 precursor, putative [Ricinus communis] |
| 4 |
Hb_004957_050 |
0.1675394052 |
- |
- |
hypothetical protein POPTR_0010s12360g [Populus trichocarpa] |
| 5 |
Hb_000580_110 |
0.1705909142 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3 [Jatropha curcas] |
| 6 |
Hb_007590_100 |
0.1719680437 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 4-like [Jatropha curcas] |
| 7 |
Hb_029223_040 |
0.1726431767 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 8 |
Hb_001621_100 |
0.1726944984 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.4 [Jatropha curcas] |
| 9 |
Hb_003052_080 |
0.1775663837 |
desease resistance |
Gene Name: PRK |
PREDICTED: uridine-cytidine kinase C [Jatropha curcas] |
| 10 |
Hb_002759_070 |
0.1786857644 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 7 [Jatropha curcas] |
| 11 |
Hb_000438_120 |
0.1870538261 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 12 |
Hb_000236_180 |
0.1913152265 |
- |
- |
serine hydroxymethyltransferase, putative [Ricinus communis] |
| 13 |
Hb_000008_060 |
0.1926663083 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_000975_170 |
0.1946205066 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 6 isoform X1 [Jatropha curcas] |
| 15 |
Hb_004032_110 |
0.1975960885 |
- |
- |
PREDICTED: putative methylesterase 14, chloroplastic [Jatropha curcas] |
| 16 |
Hb_009545_030 |
0.2001241859 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000077_310 |
0.2003831135 |
- |
- |
PREDICTED: clathrin light chain 1 [Jatropha curcas] |
| 18 |
Hb_000417_380 |
0.200460099 |
- |
- |
SecY protein transport family protein [Theobroma cacao] |
| 19 |
Hb_001546_010 |
0.2018591658 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_012707_010 |
0.2022211648 |
- |
- |
PREDICTED: uncharacterized protein C594.04c [Jatropha curcas] |