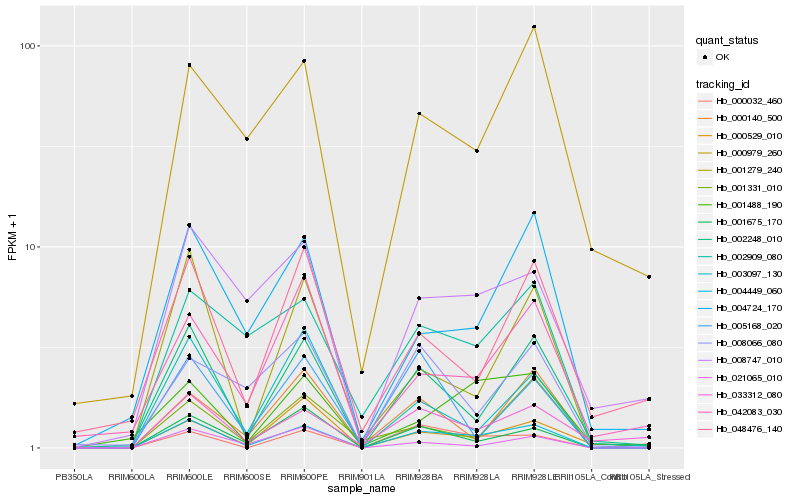
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004449_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631660 [Jatropha curcas] |
| 2 |
Hb_002248_010 |
0.1564161047 |
- |
- |
PREDICTED: uncharacterized protein LOC105124180 [Populus euphratica] |
| 3 |
Hb_000032_460 |
0.1891180738 |
- |
- |
calcineurin B, putative [Ricinus communis] |
| 4 |
Hb_001675_170 |
0.2027686107 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 5 [Malus domestica] |
| 5 |
Hb_002909_080 |
0.2051460529 |
- |
- |
inositol or phosphatidylinositol kinase, putative [Ricinus communis] |
| 6 |
Hb_001279_240 |
0.2064649288 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105633500 [Jatropha curcas] |
| 7 |
Hb_004724_170 |
0.2094653645 |
- |
- |
PREDICTED: rho GTPase-activating protein 1 [Jatropha curcas] |
| 8 |
Hb_033312_080 |
0.2103451191 |
- |
- |
aldose-1-epimerase, putative [Ricinus communis] |
| 9 |
Hb_003097_130 |
0.2131248104 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_021065_010 |
0.2169654202 |
- |
- |
PREDICTED: wall-associated receptor kinase 2-like isoform X2 [Gossypium raimondii] |
| 11 |
Hb_008747_010 |
0.2207305974 |
- |
- |
PREDICTED: protein argonaute 2-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_001488_190 |
0.2213318182 |
- |
- |
alpha-amylase, putative [Ricinus communis] |
| 13 |
Hb_008066_080 |
0.2268104712 |
- |
- |
PREDICTED: GDSL esterase/lipase 1-like [Jatropha curcas] |
| 14 |
Hb_005168_020 |
0.2276444012 |
- |
- |
PREDICTED: U-box domain-containing protein 30-like [Jatropha curcas] |
| 15 |
Hb_000979_260 |
0.2304302129 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 16 |
Hb_000140_500 |
0.2308851812 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 17 |
Hb_001331_010 |
0.2330869671 |
- |
- |
hypothetical protein CISIN_1g039303mg, partial [Citrus sinensis] |
| 18 |
Hb_042083_030 |
0.2347198826 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_000529_010 |
0.2363095546 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 20 |
Hb_048476_140 |
0.2385746524 |
- |
- |
PREDICTED: dynamin-related protein 1C [Jatropha curcas] |