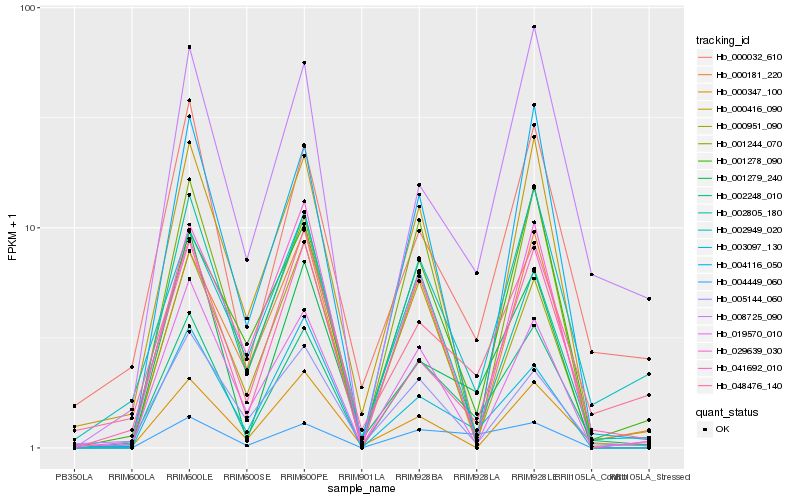
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002248_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105124180 [Populus euphratica] |
| 2 |
Hb_041692_010 |
0.1501917821 |
- |
- |
annexin [Manihot esculenta] |
| 3 |
Hb_000032_610 |
0.1522567092 |
- |
- |
PREDICTED: 3-oxoacyl-[acyl-carrier-protein] synthase I, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_048476_140 |
0.156242176 |
- |
- |
PREDICTED: dynamin-related protein 1C [Jatropha curcas] |
| 5 |
Hb_004449_060 |
0.1564161047 |
- |
- |
PREDICTED: uncharacterized protein LOC105631660 [Jatropha curcas] |
| 6 |
Hb_019570_010 |
0.159039592 |
- |
- |
lyase, putative [Ricinus communis] |
| 7 |
Hb_000416_090 |
0.1615255063 |
- |
- |
Aquaporin NIP1.1, putative [Ricinus communis] |
| 8 |
Hb_001279_240 |
0.1648127484 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105633500 [Jatropha curcas] |
| 9 |
Hb_004116_050 |
0.1654081789 |
- |
- |
PREDICTED: cytochrome P450 90A1 [Jatropha curcas] |
| 10 |
Hb_008725_090 |
0.1654137134 |
- |
- |
PREDICTED: uncharacterized protein LOC105642581 [Jatropha curcas] |
| 11 |
Hb_000951_090 |
0.1660710037 |
- |
- |
Oligopeptide transporter, putative [Ricinus communis] |
| 12 |
Hb_001244_070 |
0.1675256689 |
- |
- |
hypothetical protein POPTR_0004s13470g [Populus trichocarpa] |
| 13 |
Hb_005144_060 |
0.1681105344 |
- |
- |
Disease resistance family protein / LRR family protein, putative [Theobroma cacao] |
| 14 |
Hb_001278_090 |
0.16941649 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 15 |
Hb_002949_020 |
0.1697327736 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0002s23650g [Populus trichocarpa] |
| 16 |
Hb_000181_220 |
0.1699430406 |
- |
- |
PREDICTED: MATE efflux family protein LAL5-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_003097_130 |
0.1714991368 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002805_180 |
0.1721502795 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_029639_030 |
0.17265936 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 2-like [Jatropha curcas] |
| 20 |
Hb_000347_100 |
0.172930603 |
transcription factor |
TF Family: C3H |
hypothetical protein RCOM_0684740 [Ricinus communis] |