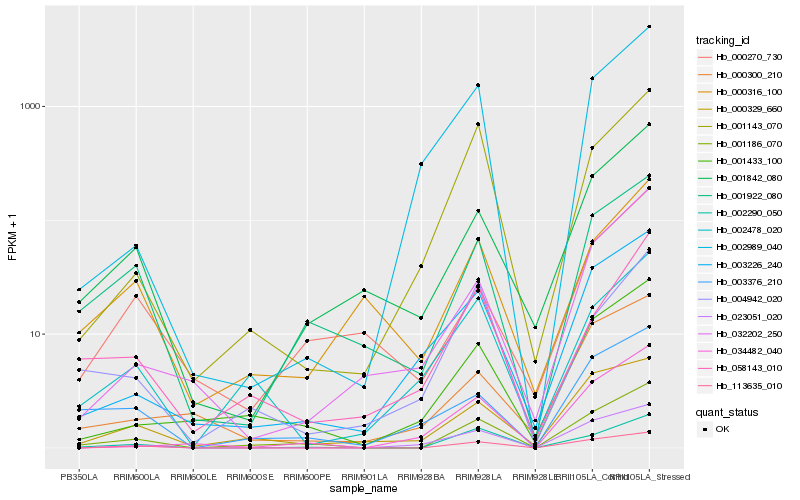
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001186_070 |
0.0 |
- |
- |
PREDICTED: F-box protein FBW2-like [Gossypium raimondii] |
| 2 |
Hb_002290_050 |
0.142215708 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001922_080 |
0.1554124689 |
- |
- |
Tellurite resistance protein tehA, putative [Ricinus communis] |
| 4 |
Hb_113635_010 |
0.1570495708 |
- |
- |
- |
| 5 |
Hb_001433_100 |
0.1593314761 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000329_660 |
0.1739377413 |
- |
- |
PREDICTED: carotenoid cleavage dioxygenase 7, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001842_080 |
0.1758555057 |
- |
- |
- |
| 8 |
Hb_003226_240 |
0.1853014851 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 9 |
Hb_002478_020 |
0.1858955966 |
- |
- |
hypothetical protein POPTR_0010s19120g [Populus trichocarpa] |
| 10 |
Hb_001143_070 |
0.1903638525 |
- |
- |
Acyl-CoA N-acyltransferases (NAT) superfamily protein isoform 1 [Theobroma cacao] |
| 11 |
Hb_003376_210 |
0.1911151609 |
- |
- |
- |
| 12 |
Hb_058143_010 |
0.197258397 |
- |
- |
hypothetical protein JCGZ_01992 [Jatropha curcas] |
| 13 |
Hb_004942_020 |
0.1991018827 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 14 |
Hb_034482_040 |
0.1994036859 |
- |
- |
aspartate kinase, partial [Francisella tularensis] |
| 15 |
Hb_023051_020 |
0.1996373907 |
- |
- |
- |
| 16 |
Hb_000270_730 |
0.2035857 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2-23 kDa-like [Jatropha curcas] |
| 17 |
Hb_032202_250 |
0.2044623673 |
- |
- |
PREDICTED: F-box protein PP2-A13 [Jatropha curcas] |
| 18 |
Hb_000300_210 |
0.2058715197 |
- |
- |
gd2b, putative [Ricinus communis] |
| 19 |
Hb_002989_040 |
0.2062912673 |
- |
- |
hypothetical protein 1 [Hevea brasiliensis] |
| 20 |
Hb_000316_100 |
0.2065908125 |
- |
- |
protein with unknown function [Ricinus communis] |