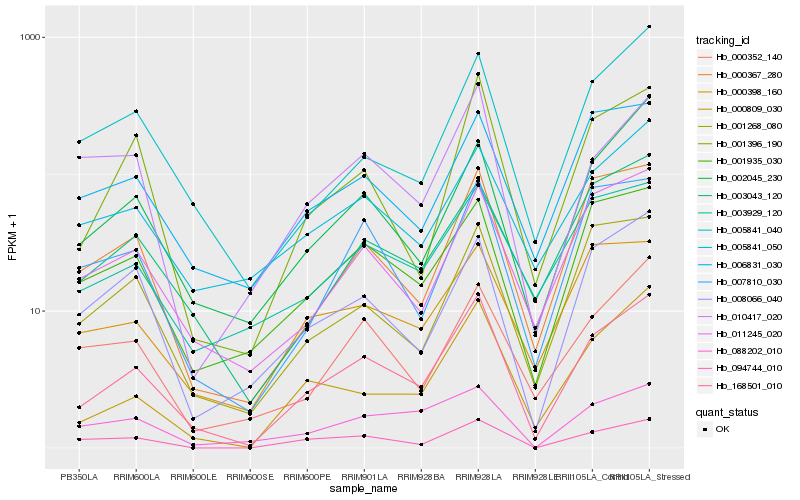
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_168501_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_000367_280 |
0.1344543069 |
- |
- |
PREDICTED: flavin-containing monooxygenase FMO GS-OX-like 9 [Jatropha curcas] |
| 3 |
Hb_001396_190 |
0.1349635911 |
- |
- |
PREDICTED: uncharacterized protein LOC105123025 [Populus euphratica] |
| 4 |
Hb_000809_030 |
0.1353850014 |
desease resistance |
Gene Name: RPW8 |
PREDICTED: probable disease resistance protein At5g66900 [Prunus mume] |
| 5 |
Hb_001935_030 |
0.1381388754 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_011245_020 |
0.1391650317 |
- |
- |
Ferrochelatase [Medicago truncatula] |
| 7 |
Hb_002045_230 |
0.1432394127 |
- |
- |
Ammonium transporter 2 [Theobroma cacao] |
| 8 |
Hb_094744_010 |
0.1461116341 |
- |
- |
PREDICTED: disease resistance protein At4g27190-like [Populus euphratica] |
| 9 |
Hb_001268_080 |
0.1466208325 |
- |
- |
catalytic, putative [Ricinus communis] |
| 10 |
Hb_007810_030 |
0.1469752905 |
- |
- |
PREDICTED: uncharacterized protein LOC105647693 [Jatropha curcas] |
| 11 |
Hb_003043_120 |
0.1497736016 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_008066_040 |
0.1521433521 |
transcription factor |
TF Family: M-type |
PREDICTED: agamous-like MADS-box protein AGL62 [Jatropha curcas] |
| 13 |
Hb_006831_030 |
0.1557366559 |
- |
- |
PREDICTED: ras-related protein RABB1c [Fragaria vesca subsp. vesca] |
| 14 |
Hb_003929_120 |
0.1570320906 |
- |
- |
PREDICTED: 50S ribosomal protein L3-2, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000398_160 |
0.1572147271 |
- |
- |
GDP-mannose transporter, putative [Ricinus communis] |
| 16 |
Hb_005841_040 |
0.1575626846 |
- |
- |
stress-induced hydrophobic peptide 1 [Hevea brasiliensis] |
| 17 |
Hb_005841_050 |
0.1594077673 |
- |
- |
PREDICTED: F-box protein SKIP1-like [Jatropha curcas] |
| 18 |
Hb_010417_020 |
0.1606418928 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 19 |
Hb_000352_140 |
0.1627939966 |
- |
- |
- |
| 20 |
Hb_088202_010 |
0.1664742118 |
- |
- |
hypothetical protein JCGZ_13241 [Jatropha curcas] |