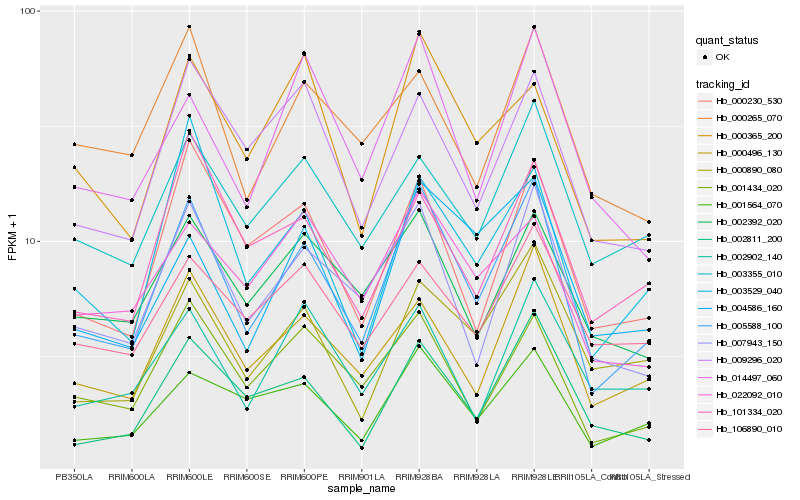
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005588_100 |
0.0 |
- |
- |
protein phosphatase, putative [Ricinus communis] |
| 2 |
Hb_007943_150 |
0.1071155864 |
- |
- |
PREDICTED: proteinaceous RNase P 1, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 3 |
Hb_000496_130 |
0.1122199164 |
- |
- |
PREDICTED: fructokinase-1 [Jatropha curcas] |
| 4 |
Hb_001564_070 |
0.1140220128 |
- |
- |
PREDICTED: G patch domain-containing protein TGH [Jatropha curcas] |
| 5 |
Hb_002811_200 |
0.1219354846 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14-like [Jatropha curcas] |
| 6 |
Hb_001434_020 |
0.1237252968 |
- |
- |
hypothetical protein RCOM_0841800 [Ricinus communis] |
| 7 |
Hb_000230_530 |
0.1285403079 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000890_080 |
0.1289886177 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 9 |
Hb_003529_040 |
0.1352500112 |
- |
- |
PREDICTED: ATP phosphoribosyltransferase 2, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_004586_160 |
0.1367748392 |
- |
- |
PREDICTED: glucose-6-phosphate isomerase 1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_002392_020 |
0.1375482052 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase [Jatropha curcas] |
| 12 |
Hb_003355_010 |
0.1377001846 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 13 |
Hb_014497_060 |
0.1388947815 |
- |
- |
phosphofructokinase, putative [Ricinus communis] |
| 14 |
Hb_009296_020 |
0.1393248444 |
desease resistance |
Gene Name: ATP-synt_ab_N |
PREDICTED: ATP synthase subunit beta, mitochondrial-like [Jatropha curcas] |
| 15 |
Hb_101334_020 |
0.1424278621 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C2A9.03-like [Jatropha curcas] |
| 16 |
Hb_000365_200 |
0.1445491146 |
- |
- |
PREDICTED: 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [Jatropha curcas] |
| 17 |
Hb_000265_070 |
0.1452581079 |
- |
- |
glucose-1-phosphate denylyltransferase small subunit [Hevea brasiliensis] |
| 18 |
Hb_022092_010 |
0.1463130363 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3-like [Jatropha curcas] |
| 19 |
Hb_002902_140 |
0.1467820172 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 9 [Jatropha curcas] |
| 20 |
Hb_106890_010 |
0.1488438977 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |