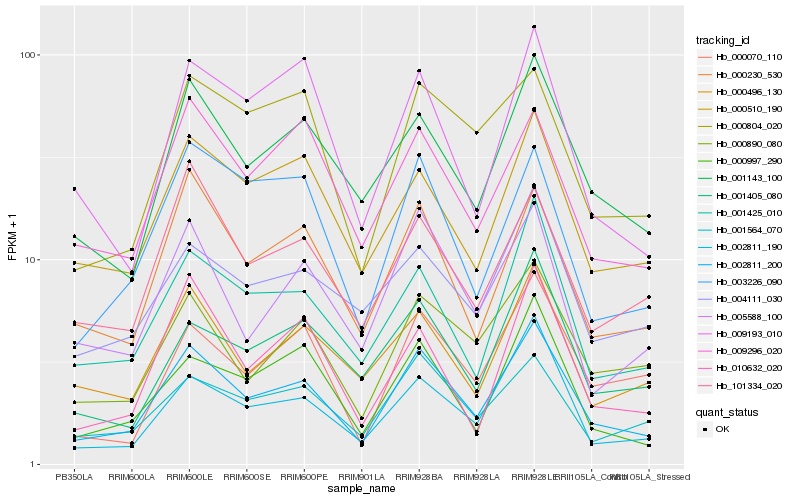
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002811_200 |
0.0 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14-like [Jatropha curcas] |
| 2 |
Hb_001143_100 |
0.1075909035 |
- |
- |
PREDICTED: delta-aminolevulinic acid dehydratase, chloroplastic [Populus euphratica] |
| 3 |
Hb_001425_010 |
0.1105311346 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 4 |
Hb_000230_530 |
0.1115342731 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002811_190 |
0.1150085352 |
- |
- |
tRNA pseudouridine synthase d, putative [Ricinus communis] |
| 6 |
Hb_000890_080 |
0.1169210386 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 7 |
Hb_000997_290 |
0.1198597566 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 8 |
Hb_001564_070 |
0.1209642313 |
- |
- |
PREDICTED: G patch domain-containing protein TGH [Jatropha curcas] |
| 9 |
Hb_000510_190 |
0.1212802928 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 10 |
Hb_005588_100 |
0.1219354846 |
- |
- |
protein phosphatase, putative [Ricinus communis] |
| 11 |
Hb_000496_130 |
0.1231833611 |
- |
- |
PREDICTED: fructokinase-1 [Jatropha curcas] |
| 12 |
Hb_003226_090 |
0.12346401 |
- |
- |
PREDICTED: bifunctional epoxide hydrolase 2 [Jatropha curcas] |
| 13 |
Hb_009296_020 |
0.1294094567 |
desease resistance |
Gene Name: ATP-synt_ab_N |
PREDICTED: ATP synthase subunit beta, mitochondrial-like [Jatropha curcas] |
| 14 |
Hb_001405_080 |
0.1306888865 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 15 |
Hb_101334_020 |
0.1312370349 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C2A9.03-like [Jatropha curcas] |
| 16 |
Hb_004111_030 |
0.1315759383 |
- |
- |
PREDICTED: DNA gyrase subunit B, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 17 |
Hb_000070_110 |
0.1316021891 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 18 |
Hb_010632_020 |
0.1318580481 |
- |
- |
hypothetical protein POPTR_0006s185902g, partial [Populus trichocarpa] |
| 19 |
Hb_009193_010 |
0.1319117525 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 20 |
Hb_000804_020 |
0.1327385108 |
- |
- |
PREDICTED: aconitate hydratase, cytoplasmic [Jatropha curcas] |