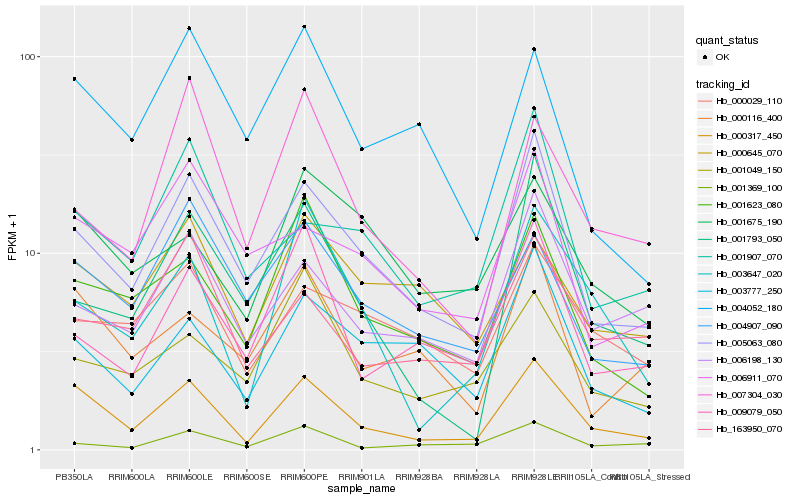
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000317_450 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647032 isoform X1 [Jatropha curcas] |
| 2 |
Hb_005063_080 |
0.1448775265 |
- |
- |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_003647_020 |
0.1751044811 |
- |
- |
hypothetical protein JCGZ_25095 [Jatropha curcas] |
| 4 |
Hb_001623_080 |
0.1783486872 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 5 |
Hb_000116_400 |
0.1853740829 |
- |
- |
protein with unknown function [Ricinus communis] |
| 6 |
Hb_003777_250 |
0.1956719533 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming]-like [Jatropha curcas] |
| 7 |
Hb_006198_130 |
0.1965660552 |
- |
- |
PREDICTED: uncharacterized protein LOC105644406 [Jatropha curcas] |
| 8 |
Hb_004907_090 |
0.1996605399 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_009079_050 |
0.2024069635 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 10 |
Hb_001907_070 |
0.2029830529 |
- |
- |
50S ribosomal protein L1p, putative [Ricinus communis] |
| 11 |
Hb_001793_050 |
0.2042252905 |
- |
- |
PREDICTED: uncharacterized protein LOC105632663 [Jatropha curcas] |
| 12 |
Hb_001675_190 |
0.2062453752 |
- |
- |
PREDICTED: probable plastidic glucose transporter 3 [Jatropha curcas] |
| 13 |
Hb_004052_180 |
0.2077590225 |
- |
- |
PREDICTED: protein COBRA-like [Jatropha curcas] |
| 14 |
Hb_163950_070 |
0.2082114051 |
- |
- |
arginine/serine-rich splicing factor, putative [Ricinus communis] |
| 15 |
Hb_001049_150 |
0.2092703565 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 11 [Jatropha curcas] |
| 16 |
Hb_000645_070 |
0.2096723537 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_001369_100 |
0.2096739624 |
- |
- |
hypothetical protein JCGZ_26280 [Jatropha curcas] |
| 18 |
Hb_007304_030 |
0.2101630174 |
- |
- |
PREDICTED: uncharacterized protein LOC105635926 isoform X2 [Jatropha curcas] |
| 19 |
Hb_006911_070 |
0.211058096 |
- |
- |
50S ribosomal protein L1p, putative [Ricinus communis] |
| 20 |
Hb_000029_110 |
0.2115186219 |
- |
- |
conserved hypothetical protein [Ricinus communis] |