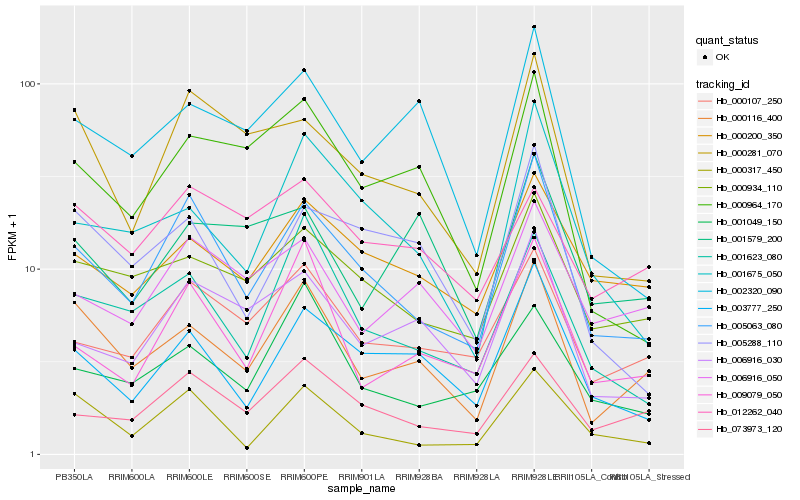
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000116_400 |
0.0 |
- |
- |
protein with unknown function [Ricinus communis] |
| 2 |
Hb_000934_110 |
0.1519995367 |
- |
- |
PREDICTED: protein trichome berefringence-like 7 [Jatropha curcas] |
| 3 |
Hb_005063_080 |
0.1576900344 |
- |
- |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_001623_080 |
0.1637676426 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 5 |
Hb_002320_090 |
0.166464524 |
- |
- |
PREDICTED: endoglucanase 25-like [Jatropha curcas] |
| 6 |
Hb_000200_350 |
0.1709587762 |
- |
- |
PREDICTED: uncharacterized protein LOC105636929 [Jatropha curcas] |
| 7 |
Hb_000964_170 |
0.171814763 |
- |
- |
Cellulose synthase A catalytic subunit 6 [UDP-forming], putative [Ricinus communis] |
| 8 |
Hb_003777_250 |
0.1734254702 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming]-like [Jatropha curcas] |
| 9 |
Hb_000107_250 |
0.1757651226 |
- |
- |
voltage-gated clc-type chloride channel, putative [Ricinus communis] |
| 10 |
Hb_006916_050 |
0.1776925137 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 11 |
Hb_009079_050 |
0.1789222502 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 12 |
Hb_073973_120 |
0.1789321604 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_000281_070 |
0.179643456 |
- |
- |
rubisco subunit binding-protein beta subunit, rubb, putative [Ricinus communis] |
| 14 |
Hb_005288_110 |
0.1801665222 |
- |
- |
PREDICTED: uncharacterized protein LOC105647752 [Jatropha curcas] |
| 15 |
Hb_001579_200 |
0.1833199638 |
- |
- |
nucleoredoxin, putative [Ricinus communis] |
| 16 |
Hb_000317_450 |
0.1853740829 |
- |
- |
PREDICTED: uncharacterized protein LOC105647032 isoform X1 [Jatropha curcas] |
| 17 |
Hb_006916_030 |
0.1857010536 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 18 |
Hb_001049_150 |
0.1861594505 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 11 [Jatropha curcas] |
| 19 |
Hb_012262_040 |
0.1867614102 |
- |
- |
PREDICTED: uncharacterized protein LOC105633697 [Jatropha curcas] |
| 20 |
Hb_001675_050 |
0.1870478199 |
- |
- |
PREDICTED: uncharacterized protein LOC105635000 [Jatropha curcas] |