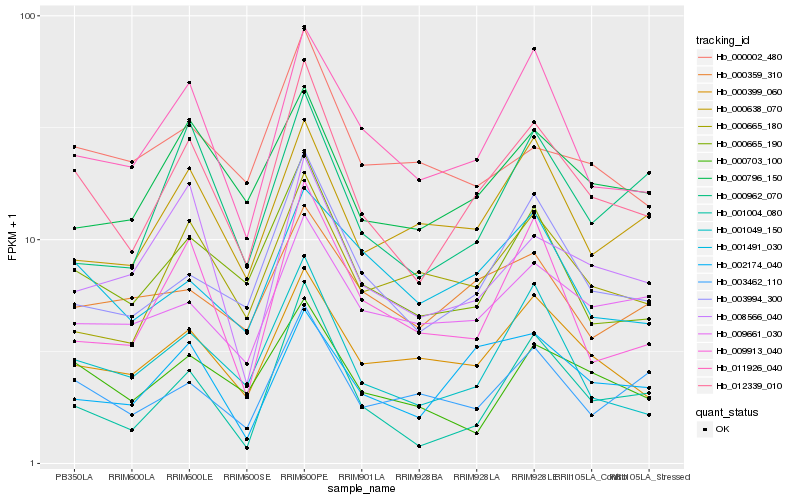
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012339_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000665_190 |
0.1132548231 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_011926_040 |
0.1191081669 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003994_300 |
0.1195057895 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001049_150 |
0.1262793566 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 11 [Jatropha curcas] |
| 6 |
Hb_000399_060 |
0.1280469845 |
- |
- |
PREDICTED: uncharacterized protein LOC105649658 [Jatropha curcas] |
| 7 |
Hb_002174_040 |
0.1327443124 |
- |
- |
DNAJ heat shock family protein [Theobroma cacao] |
| 8 |
Hb_000796_150 |
0.1354446567 |
- |
- |
PREDICTED: uncharacterized protein LOC105136996 isoform X1 [Populus euphratica] |
| 9 |
Hb_000703_100 |
0.1393850358 |
- |
- |
PREDICTED: bifunctional riboflavin kinase/FMN phosphatase-like [Jatropha curcas] |
| 10 |
Hb_001004_080 |
0.1415147949 |
- |
- |
hypothetical protein POPTR_0014s14130g [Populus trichocarpa] |
| 11 |
Hb_008566_040 |
0.141853427 |
- |
- |
PREDICTED: uncharacterized protein LOC105647590 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003462_110 |
0.1419927076 |
- |
- |
hevamine-A precursor, putative [Ricinus communis] |
| 13 |
Hb_009661_030 |
0.1443385864 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like [Jatropha curcas] |
| 14 |
Hb_000962_070 |
0.1446437758 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 13, chloroplastic isoform X2 [Jatropha curcas] |
| 15 |
Hb_000638_070 |
0.1465594286 |
- |
- |
PREDICTED: protein trichome birefringence-like 14 [Jatropha curcas] |
| 16 |
Hb_000665_180 |
0.1485443551 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |
| 17 |
Hb_001491_030 |
0.1490268788 |
- |
- |
PREDICTED: uncharacterized protein LOC105649804 isoform X1 [Jatropha curcas] |
| 18 |
Hb_009913_040 |
0.1504441881 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 19 |
Hb_000359_310 |
0.1545625274 |
- |
- |
hypothetical protein JCGZ_03934 [Jatropha curcas] |
| 20 |
Hb_000002_480 |
0.1572649954 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X2 [Jatropha curcas] |