| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
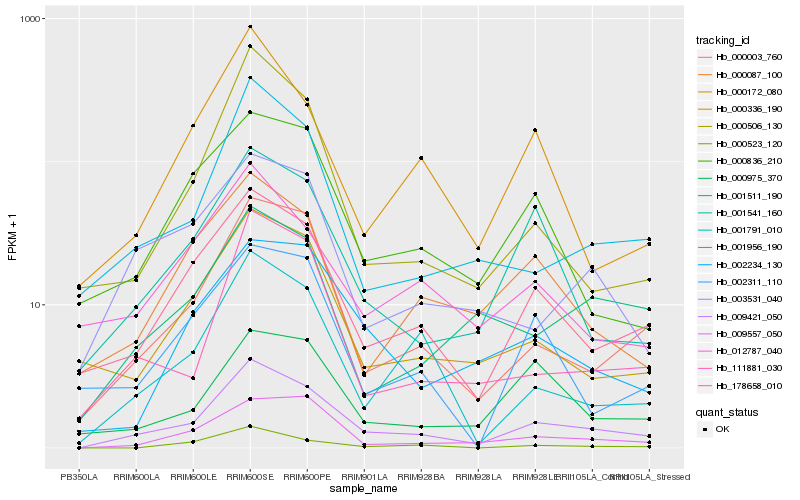
Hb_009421_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647037 [Jatropha curcas] |
| 2 |
Hb_178658_010 |
0.1383129788 |
- |
- |
PREDICTED: uncharacterized protein LOC105641396 [Jatropha curcas] |
| 3 |
Hb_000003_760 |
0.1533353811 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA26-like [Jatropha curcas] |
| 4 |
Hb_009557_050 |
0.1707107178 |
transcription factor |
TF Family: C2H2 |
zinc finger family protein [Populus trichocarpa] |
| 5 |
Hb_001541_160 |
0.1736897769 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 6 |
Hb_001956_190 |
0.1738352604 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 7 |
Hb_000336_190 |
0.1755370724 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Gossypium raimondii] |
| 8 |
Hb_001791_010 |
0.1819632387 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g63710 [Jatropha curcas] |
| 9 |
Hb_111881_030 |
0.1835816644 |
- |
- |
PREDICTED: uncharacterized protein LOC105645255 [Jatropha curcas] |
| 10 |
Hb_002234_130 |
0.1870637479 |
- |
- |
ATPP2-A13, putative [Ricinus communis] |
| 11 |
Hb_000836_210 |
0.1889396074 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 12 |
Hb_000087_100 |
0.1894694093 |
- |
- |
PREDICTED: cellulose synthase-like protein D3 [Jatropha curcas] |
| 13 |
Hb_012787_040 |
0.1945857356 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000172_080 |
0.1955139842 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |
| 15 |
Hb_003531_040 |
0.1960492158 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 16 |
Hb_000975_370 |
0.1965154277 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB59 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000506_130 |
0.196840293 |
- |
- |
DGCR14-related family protein [Populus trichocarpa] |
| 18 |
Hb_001511_190 |
0.1971409206 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 19 |
Hb_002311_110 |
0.2007158096 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000523_120 |
0.20077104 |
- |
- |
PREDICTED: butyrate--CoA ligase AAE11, peroxisomal-like [Jatropha curcas] |